
The exposome-diet-epigenome axis in inflammatory bowel diseases—a narrative review
Introduction
Inflammatory bowel disease (IBD) is a progressive immune-mediated chronic gastrointestinal (GI) disease, including Crohn’s disease (CD) and ulcerative colitis (UC), and is exemplified by a persistent relapsing-remitting inflammation profile (1). IBD is a complex condition involving multiple factors, including genetics, environment, and gut microbiota (2,3). The most common theory is that it is caused by a faulty immune response in the gut, which leads to an impaired intestinal barrier. Factors related to lifestyles, such as diet, physical activity, medication use, sleep, anxiety, and pollution, are essential in evaluating the risk of developing IBD (4). The incidence of IBD is increasing worldwide, especially in recently industrialized areas worldwide, such as South America, Africa, and Asia (1,5). More than 3 million people have been diagnosed with IBD. Medical care costs are high to the public health care systems, and several patients become less productive due to sickness, leading to disability and premature retirement (6,7).
The Montreal Classification for CD is a worldwide system used to characterize CD patients (8). According to this classification, CD is classified into three main categories: age at diagnosis, location of disease, and disease behavior. The age-at-diagnosis category divides patients into pediatric (<17 years old) and adult (≥17 years old) groups (8). The location of the disease is classified according to the inflamed area in the GI tract and includes three subcategories: L1 (terminal ileum), L2 (colon), L3 (ileocolon), and L4 (upper GI). Disease behavior is classified into three subcategories: B1 (non-penetrating and non-stricturing disease), B2 (stricturing), and B3 (penetrating). Furthermore, the Montreal Classification also includes additional categories to classify disease behavior based on the presence or absence of perianal disease, which would be indicated by a ‘p’ appended to behavior categories (B1p, B2p, and B3p). This classification system is useful in predicting disease outcomes and guiding treatment decisions in patients with CD (8).
Environmental factors are important in the genesis of IBD manifestations. A few decades ago, with the increase in IBD incidence and research advances about the metabolomic, new pathways were explored to better understand how environmental factors are associated with the disease (9). Epigenetic regulation is an important explanation for the influence of environmental factors on the development of IBD (10). The epigenome, which refers to the chemical modifications of DNA, can directly influence different processes, such as transcription, replication, recombination, repair, histone methylation, and modification (11).
The crucial role of epigenetics and the more accepted concept of the “epigenome” has motivated many researchers to investigate different approaches. The epigenome plays a fundamental role in controlling gene expression and regulating critical biological processes such as embryonic development, cellular differentiation, and response to environmental stimuli (4,9-12). Diet is one of the most critical environmental factors in the development of IBD (9). Nutrients can, directly and indirectly, affect the epigenome, and diet can alter the microbiome, leading to epigenetic modifications (10). Although we do not know the exact cause of remission for IBD, exploring the contribution of epigenetics, especially diet, may help develop future therapeutic approaches to improve the outcome of the disease (9,10,13). In this review, we discuss the role of diet in modulating the microbiome and influencing epigenetic changes through the exposome-diet-epigenome axis. We could hence favor a future therapeutic approach to improve the outcome of the disease. We present this article in accordance with the Narrative Review reporting checklist (available at https://dmr.amegroups.com/article/view/10.21037/dmr-22-85/rc)
Methods
To interpret the evidence on the association of environmental factors and endogenous factors in individuals with IBD and create a connection between these factors, we performed a thorough literature review. We selected articles published from 1942 to 2023 by searching the MEDLINE database at the NIH National Library of Medicine PubMed (Table 1). In this database, we selected articles indexed in English. Editorial comments, technical reports, abstracts published in scientific events, and case studies were excluded.
Table 1
| Items | Specification |
|---|---|
| Date of search | November 2022 to March 2023 |
| Databases and other sources searched | NIH National Library of Medicine PubMed, MEDLINE database |
| Search terms used | Inflammatory bowel disease; diet; exposome; epigenetic; Crohn disease; ulcerative colitis; microbiota; miRNAs; DNA modifications |
| Timeframe | 1942 to 2023 |
| Inclusion and exclusion criteria | Only English literature were included. |
NIH, National Institutes of Health.
Genome in IBDs
IBD is a complex and multifactorial disorder influenced by various factors, including genetics and the environment (14). The genome is a significant risk factor for disease development (11-14). IBD among relatives and twins reiterates its importance (11,12). Both main IBD have a genetic component, but the heritability factor is relatively higher in CD than in UC (9).
Since the 1980s, IBD has been considered a polygenic disease, and several epidemiological studies have found multiple susceptibility loci associated with the occurrence of the disease (13,14). Genome-Wide Association Studies (GWAS) have successfully identified about 240 loci contributing to IBD susceptibility (15). Several loci showed specific mutations resulting in loss-of-function immune pathway variants that generally respond to the host microbiome for intestinal homeostasis (16-18). For example, the Nucleotide-binding oligomerization domain-containing protein 2 (NOD2, also known as CARD15) was the first gene identified through genome studies for CD susceptibility (19,20). NOD2 discovery suggested that pattern recognition receptors and innate immunity have an important role in physio pathogenesis (13). Another important cellular-stress mechanism with polymorphisms related to IBD is autophagy. GWAS analysis also found the gene ATG16L1 (autophagy-related 16-like 1) and IRGM (immunity-related GTPase family protein) linked to IBD (15,17). The role of the inflammatory signaling pathways, such as IL 23-driven T helper cell responses, also has been described in IBD (12,13,16,21). IL10 locus is associated with UC and CD with potential therapeutic functionality (22). Two meta-analyses have revealed 47 UC susceptibility loci, and 71 were shown for CD (22,23). The analysis emphasized the role of impaired barrier function in UC etiology (23). Indeed, some genetic variants that control epithelial barrier functions have been reported to be associated with IBD, such as TTC7A, which maintains apicobasal epithelial polarity, and EPCAM and SPINT2 epithelial adhesion genes (18).
Other autoimmune and immune-mediated diseases such as rheumatoid arthritis, psoriasis, multiple sclerosis, type 1 diabetes, and celiac disease commonly share some genetic locus (21,22). For example, ankylosing spondylitis and IBD are associated with IL23R and TNFSF15 (22). Celiac disease shares four risk loci with CD (PTPN2, IL18RAP, TAGAP, and PUS10) (24). González-Serna and coauthors showed four new loci share between systemic sclerosis and CD (IL12RB2, IRF1/SLC22A5, STAT3, and an intergenic locus at 6p21.31) (25).
Despite the success in identifying several loci and replicable genome studies associations, all these genetic factors explain a modest fraction of phenotypic variance of 08% for UC and 13% for CD, demonstrating the complexity of IBD disorders (26). According to Fofanova et al. [2016], the phenotypic concordance in twin studies is between 27–56% for CD and less than 20% for UC (9). That is, the genetic predisposition alone is inefficient in determining the development or not of IBD (9). Furthermore, the increase in the incidence of chronic-immune diseases, besides IBD, demonstrates that other environmental factors influence disease pathogenesis in addition to genetic factors (25,27).
Environmental factors “exposome”
Chronic immune-mediated diseases such as IBD are emerging whiting industrialization (14,28). Increasing evidence suggests that environmental factors have an important role in IBD incidence and outcome (4). IBD is considered a disease of the industrialized world (29,30). Several studies have shown epidemiological evidence of the association of environmental factors, such as urbanization, westernized diet, air pollution, food chemical additives, smoking, psychological factors, sleep disturbances, exercise, antibiotics, oral contraceptive pills, and microplastic exposure to IBD (31-33).
However, the mechanism behind these remains unknown. These factors can be analyzed in IBD by exerting an effect on the gut microbiome. Improved sanitation and personal hygiene in developed countries have reduced infectious diseases. However, some studies have shown that better health conditions are associated with an increased risk of IBD. Research compiled by Vedamurthy and Ananthakrishnan [2019] suggests that living in a large family, crowded rural homes, having a pet at home, consuming unpasteurized milk, being in contact with Helicobacter pylori, and parasitic worm colonization are negatively associated with the prevalence of IBD (32). A hygienic environment has reduced the number and diversity of microbiota, resulting in changes in the microbiome profile. Fewer episodes of infections throughout life can result in an abnormal immune response (34).
Other external environmental factors influencing IBD consist of the maternal lifestyle and in-utero events, mode of child delivery (vaginal vs. cesarean), breastfeeding, food choices, smoking, drugs, vaccination, alcoholism, physical activity, stress, appendicectomy, vitamin D deficiency, and others factors not yet discovered until now (4,35-37). Curiously, the smoking habit has the opposite effect in UC than in CD (38-41). Smoking increases smokers’ risk of developing CD, and disease outcome is impaired (33). In contrast, smoking protects against UC and favors better disease development (38-40). In this context, several potential active mediators in smoke may be responsible for these clinical effects, including nicotine and carbon monoxide, which could modulate the autoantibodies production, inflammatory profile, and leukocyte migration. However, the precise mechanism remains unknown (38,41).
Epigenetics can be mitotically heritable changes in gene function that cannot be explained by altered DNA sequences (12). It can influence the transcriptional expression contributing to IBD by working through DNA methylation, histone alterations, or silencing by non-coding RNAs (ncRNAs) (4,42,43). How these environmental factors induce changes in the exposome of individuals needs to be better studied. Through epigenetics studies, it is possible to evaluate their influence in terms of duration, frequency, and time they occur and how they modulate IBD.
Diet in IBDs
The higher incidence rates of IBD in Westernized societies point to the potential role of diet concerning dietary patterns and specific nutrients in developing and managing the disease (44). Western diet, characterized by a high intake of saturated fat, refined sugar, and salt and low consumption of fiber, fruits, and vegetables, is associated with gut dysbiosis, resulting in differences in microbial composition and reduced microbial diversity (35).
Furthermore, our research group showed in a review that high saturated fat, sucrose, monosaccharide consumption, and long-term fast food dietary consumption are associated with CD (44). At the same time, breastfeeding and fiber might play a protective role. Regarding UC, the dietary risk factors are overconsumption of animal fat and cholesterol, sucrose, and linoleic acid. In contrast, breastfeeding and high consumption of n-3 polyunsaturated fatty acids (PUFAs) are associated with a diminished risk of UC (44). The specific components, related to a higher IBD risk, and the Westernized diet, characterized by ultra-processed food consumption, can worsen experimental gut inflammation by exploiting the gut microbiota or engaging innate immune receptors and related cellular stress signaling (45). Through genetic colocalization and Mendelian randomization techniques, researchers have identified the causal relationship between n-6 PUFAs, and the derived metabolites as a causality factor for CD (46,47). Another study has shown the protective effects of n-3 PUFAs on IBD, commonly found in foods such as fish oil, flaxseed, and other sources (46). The recommended dietary intake ratio of omega-3:omega-6 depends on the disease under consideration (48). According to Cholewski et al. [2018], the ideal consumption would be to follow the proportion of the Japanese population from 1:2 to 1:4 (48). However, some researchers suggest consumption from 1:5 to 1:10 of n-3:n-6 would already be adequate for the general population (48).
In addition to ingesting this essential nutrient, such as the PUFA ratio, evaluating the patient’s entire dietary context is necessary. Researchers have evaluated specific diets for IBD treatment. Some nutritional strategies, such as exclusive enteral nutrition (EEN) (49) and Crohn’s Disease Exclusion Diet (CDED) (50,51), have been found effective in inducing remission, at least in CD. According to the ESPEN guideline (52), an EEN is recommended as the first-line therapy to induce remission in children and adolescents with mild active CD. This nutrition therapy is based on administering a liquid nutrition formula (polymeric diet with moderate fat content) via a feeding tube, which completely excludes food intake for several weeks (49). CDED has the advantage of allowing real food and recommends a better-acceptable alternative to the exclusive use of enteral formula. However, other dietary strategies, such as the specific carbohydrate diet (SCD), modulating early life microbiome through dietary intervention in pregnancy (MELODY), anti-inflammatory diet for IBD (IBD-AID), and low FODMAP (fermentable oligosaccharides, disaccharides, monosaccharides, and polyols) diet, have shown effects in improving symptoms (49-54).
These aspects highlight the importance of intervention studies in IBD patients to evaluate clinical outcomes and quality of life.
Gut microbiome
The microbiome plays a crucial role as an environmental factor that can modulate the exposome, leading to epigenetic modifications in the host. The microbiome study is part of an important axis that directs the occurrence of IBD: the exposome-microbiome-epigenome axis (4).
Bacterial load is the best-studied component of intestinal microorganisms. Bacterial microbiota can reach 1011–1012 organisms in the GI tract per gram of luminal content in the colon (55). This colonization by microorganisms in the gut starts during the intrauterine period and may play a role in IBD within a predisposing environment (54,56,57). Influenced by the delivery, lack of breastfeeding, and early environmental exposures, gut microbiota paves the way for early-life health or IBD (56-58). According to Ramos and Papadakis [2019], the interplay between genetic predisposition and environmental influence on the microbiome may lead to inappropriate immune activation by weakening the gut barrier (33).
The gut microbiome interacts with the host symbiotically and plays important roles: digestion of substrates and nutrient production; production of vitamins and short-chain fatty acids (SCFAs); protection of mucosal homeostasis repressing the growth of harmful microorganisms; and development and regulation of the immune system (59,60).
Influenced by the microbiome and diet, SCFAs are gut microbiota-derived metabolites produced by anaerobic fermentation of indigestible fibers mainly found in fruits, vegetables, and abundance in flaxseed, chia seed, barley, oats, and other foods (2,4,60-62). The three main SCFAs are acetate, butyrate, and propionate, which exert anti-inflammatory effects and promote the integrity of epithelial barrier function (2,4,10,60-62). These metabolites play a key role in the microbiome-host interface and maintain intestinal homeostasis by supporting epithelial cell function, stimulating mucus quality and synthesis, and strengthening intestinal barrier integrity (61). For example, Akkermansia muciniphila, a human intestinal mucin-degrading bacterium, has been identified as a critical propionate-producing (63). The microorganisms: Faecalibacterium prausnitzii, Eubacterium rectale, Eubacterium hallii, Ruminococcus bromii, Roseburia spp., Lactobacillus spp., and Bifidobacterium spp., are responsible for the most significant fraction of butyrate production (64-66). Fermentation of resistant starch is thought to contribute significantly to butyrate production in the colon, so reducing these organisms would significantly reduce their production.
Furthermore, the host-associated microbiome prevents the growth of harmful microorganisms by the support the innate immune defense against pathogenic bacteria through the stimulation of antimicrobial molecules (61). Alterations in the gut microbiota are known as dysbiosis. Dysbiosis states of the microbiome might be an environmental stimulus in altering the host-mucosal defenses and triggering immune responses favoring the disease onset (66). For example, the gut of an infant with dysbiosis promotes a strong lymphocyte T helper cell (Th1) bias, which induces the immune system to become pro-inflammatory and produce the cytokines interleukin-12 (IL-12) and interferon (IFN)-γ secretion (57,67). This inflammatory condition alters the immune system, which can lead to long-term consequences such as IBD, allergies, irritable bowel syndrome (IBS), and autoimmune diseases.
Bacterial dysbiosis modifies various chemicals triggering host reactions that may affect several physiological functions such as immunity, neurobiology, metabolism, and intestinal absorption (66,68). Three main bacterial phyla are in the human gut: Firmicutes, Bacteroidetes, and Actinobacteria (66). Proteobacteria and Verrucomicrobia are also in the human gut microbiome but in lesser quantities (67,69). IBD is characterized by decreased diversity, reduced proportions of Firmicutes, and increased proportions of Proteobacteria and Actinobacteria (70). Pro-inflammatory bacterial species actions are enriched in IBD patients (Escherichia, Fusobacterium); oppositely, anti-inflammatory species (Faecalibacterium, Roseburia) are significantly reduced in IBD. Moreover, the literature has shown a lower abundance of Clostridium coccoides, Clostridium leptum, Faecalibacterium prausnitzii, and Bifidobacterium in IBD (64,71).
The Proteobacteria phylum increases in IBD patients, including Enterobacteriaceae and Gammaproteobacteria (71). Proteobacteria are anaerobic bacteria whose expansion is facultative, usually associated with dysbiosis (72). Lactobacillus, Bifidobacterium, and Faecalibacterium are gut microbiota species that may have a protective role in IBD (70).
Accordingly, there has been a surge of interest in microbiome-based drug development as a therapeutic for IBD. Yet there is still no consensus on whether dysbiosis is a cause or consequence in the context of IBD. That relationship and the therapeutic approach benefits of its treatment require further studies (34).
Diet can modulate microbiota
Depletion of commensal microbes and consequent dysbiosis can impair mucosal healing and enhanced recruitment of proinflammatory leukocytes to the lamina propria and induce chronic inflammation and colitis in genetically predisposed individuals (60). High-fat diet (HFD) modulates the gut microbiome negatively, which can impact the mucosal integrity and immune system, altering the humoral response, mediated by B lymphocytes and antibodies produced by them, and cellular response, mediated by T lymphocytes and cytokines (73). In this case, the passage of luminal materials may be related to excessive immune activation and the release of cytokines, such as IL-4, IL-6, IL-12, IL-13, TNF, IL-1B, IFN-γ, and inhibition of cytokines, such as IL-10, IL-17, and IL-22. As a result, increased tight junction permeability may lead to the additional passage of macromolecules from the intestinal lumen and increased immune activation, exacerbating the symptoms of IBD (73). Several studies of IBD-associated microbiome have proven that dietary components can alter the intestinal microbiota and may influence epigenetic changes. Thus, diet plays a crucial role in modulating the microbiome and epithelial barrier (65). Dietary fat has sparked the interest of researchers in gut barrier regulation in IBD, particularly in Western countries. Mediterranean, Indian, Japanese, and Southeast Asian diets, rich in unsaturated fatty acids and fiber, are associated with anti-inflammatory properties (74). In contrast, a Western-style diet, rich in saturated fatty acids and sugars and poor in fiber, is associated with adverse health impacts on intestinal microbiota and might also have a role in the pathogenesis of IBD (72,73,75). Numerous studies have shown that an HFD induces gut permeability alterations due to gut microbiota changes. The results of these studies reveal that HFD expands microbes involved in reducing barrier integrity, such as Oscillibacter spp. and Desulfovibrio spp. While reducing microbes that promote the intestinal barrier, such as Bifidobacterium spp., Lactobacillus spp., Clostridiales spp., Bacteriodetes spp., and Akkermansia muciniphila (76). The exact mechanisms these specific microbial species modulate intestinal permeability are still widely debated; some have been investigated and described in a recently published comprehensive review (72).
Western-style diets are also high in simple carbohydrates such as sugar. High intakes of fermentable carbohydrates (FODMAPs) such as sucrose, fructose, glucose, lactose, or polyols (mannitol, sorbitol, xylitol, and maltitol) have been shown to exceed the absorption capacity of the intestine, increasing intestinal dysbiosis and permeability (69). They may contribute to the onset or maintenance of IBD (69). Moreover, carbohydrate malabsorption, which occurs with ingesting FODMAPs mainly, results in bacterial overgrowth. Excessive carbohydrate fermentation by bacteria in the intestinal lumen produces methane gas or hydrogen, causes abdominal distension, and contributes to IBD pathogenesis (69). The low FODMAP diet was initially used for IBS patients by acting in the reduction of de symptoms (77). In this context, that diet has been tested in IBD patients to reduce abdominal pain, bloating, flatulence, and diarrhea in both CD and UC patients in remission (77).
The high exclusion of fibers for a prolonged time can lead to harmful changes in the intestinal microbiota (69). Recent randomized controlled trials have demonstrated reduced levels of Bifidobacterium due to a low FODMAP diet (78). However, it is necessary to evaluate each patient individually. For example, reduced and controlled consumption of foods with FODMAPs for a determined period has been encouraged for IBD patients with a phenotype similar to IBS, with pain and abdominal distension (77,78).
The low FODMAP dietary strategy must be well calculated since the probability of inadequate fiber intake is very high, besides the low intake of micronutrients and bioactive compounds. This scenario can lead to constipation, dysbiosis, and nutritional deficiency. Fermented fiber within the colon produces organic acids and gases, promotes bacterial diversity, preserves mucosal barriers, and prompts the production of a large amount of SCFA (acetate, propionate, and butyrate). Non-digestible carbohydrates derived from dietary fibers and endogenous products, such as mucin, are substrates for bacterial fermentation (77).
Although acetate production pathways are widely distributed among bacterial groups, the production of propionate, butyrate, and lactate appears more highly substrate-specific (61). IBD patients show a decrease in butyrate-producing bacterial species and reduced expression of butyrate transporters. SCFA positively modulates intestinal homeostasis and reduces inflammation, which is why low fiber intake is also associated with an increased incidence of IBD (69).
Other diet protocols, like a gluten-free diet, have seemed to be used by IBD patients to treat symptoms (78,79). However, changes in the intestinal microbiota caused by the lack of grain-based foods, such as lower concentrations of Bifidobacterium spp. and Lactobacillus spp., can lead to a reduction in SCFA production, whose role is essential for several functions in the body, such as participation in our metabolism, production of anti-inflammatory mediators, and intestinal health (79).
Another food component with an important impact on the microbiota is food additives. There is scientific evidence of the adverse effects of food additives in ultra-processed foods on the microbiota (65). For example, non-nutritive sweeteners (NNSs) like saccharin, aspartame, acesulfame-K, and sucralose have been shown to significantly impact bacterial growth both in vitro and in vivo. They may alter gut microbiota composition (80). Dietary emulsifiers alter human microbiota composition and ex vivo gene expression, enhancing intestinal inflammation (81). Thus, maltodextrins commonly used in producing candies, sports energy products, and soft drinks alter the intestinal microbiota and epithelial cells, reducing mucus production and exacerbating intestinal inflammation (82,83). Like NNSs, other additives typically used to prepare ultra-processed foods can alter the gut microbiota. Xenobiotics like an organochlorine pesticide, glyphosate, and perfluorooctanesulfonic acid can negatively modulate the composition and functions of the microbiome and epithelial barrier and may even influence epigenetic changes in the gut (84,85).
Other types of diet used by IBD patients (additionally information in Table 2) that seem to alter the microbiota include the lactose-free diet, SCD, Mediterranean diet, the IBD-AID, the gluten-free diet, as well as the consumption of prebiotic foods (such as fruits and vegetables) and probiotic foods (such as yogurt, pickles, fermented foods like bread, kombucha, kefir, and also in capsules with lyophilized bacteria). However, there is no clear recommendation for using a specific diet for all patients with IBD (52). Thus, more studies must evaluate how foods modulate the microbiota, leading to a favorable outcome or not for IBD patients. In this context, plus diet, some researchers presented other environmental factors that can contribute to IBD and microbiota modulation (Table 2).
Table 2
| Environmental factors | First author, year | Main finding summary |
|---|---|---|
| Diet (pattern modifications) | Li et al., 2020 (36) | In this systematic review and meta-analysis, Li et al. aimed to investigate the association between a pre-illness Western dietary pattern and the risk of developing IBD. The authors analyzed data from seven studies that met the inclusion criteria, involving 838,413 participants. Their findings suggest that a pre-illness Western dietary pattern may be associated with an increased risk of developing IBD, particularly CD. The study highlights the importance of nutritional interventions for individuals at high risk of developing IBD and the potential role of a healthier diet in preventing the onset of this condition |
| Vasseur et al., 2021 (86) | The study investigated the associations between dietary patterns and ultra-processed food intake with the risk of developing IBD using data from the NutriNet-Santé cohort. The study results showed that a higher ultra-processed food intake was associated with an increased risk of developing IBD. In contrast, higher adherence to the “Mediterranean-like” dietary pattern was associated with a decreased risk of IBD. These findings suggest that nutritional interventions, such as reducing the intake of ultra-processed foods and promoting a Mediterranean-like dietary pattern, may be beneficial for preventing IBD | |
| Green et al., 2019 (87) | In this review article published in Nutrients in 2019, Green et al. discuss dietary therapy’s current status and future potential for IBD. The authors highlight the various nutritional approaches studied for IBD, including exclusive enteral nutrition, specific carbohydrate diet, MD, and low FODMAP diet. They also discuss how diet may affect IBD pathophysiology, such as modulating the gut microbiome, reducing inflammation, and restoring gut barrier function. The authors conclude that while the evidence for specific diets in IBD treatment is still limited, dietary therapy should be considered an essential adjunct to conventional medical therapies for IBD. Further research is needed to identify optimal nutritional strategies for managing IBD | |
| Diet (FODMAPs) | Gibson, 2017 (88) | The study suggests a low FODMAP diet can effectively improve IBS-like symptoms in patients with quiescent IBD. However, this diet should be carefully supervised to prevent nutritional risks, given the common occurrence of undernutrition in this patient population |
| Zhan et al., 2018 (89) | This study presents a meta-analysis and systematic review of studies investigating the effectiveness of a low FODMAP diet in treating patients with IBD. The results indicate that a low FODMAP diet can effectively improve gastrointestinal symptoms in patients with quiescent IBD. However, more well-designed and long-term studies are needed to confirm these findings | |
| Simões et al., 2022 (90) | This paper reviews the potential therapeutic use of a low FODMAP diet for IBD and its impact on gut microbiota. The review suggests that the low FODMAP diet may improve clinical outcomes in managing IBD, but concerns about its adequacy and effects on gut microbiota exist. The article emphasizes the importance of individualizing and monitoring the diet to avoid negative consequences and the need for further studies to understand better the relationship between diet, gut microbiota, and IBD | |
| Diet (exclusion diet) | Sigall-Boneh et al., 2014 (51) | The main finding of this study is that a structured diet combining partial enteral nutrition with the exclusion of specific dietary components leads to high remission rates in children and young adults with mild-to-moderate CD. Remission was achieved in 70.2% of patients, with normalization of previously elevated CRP in 70% of patients in remission. The results suggest that this dietary therapy approach may effectively manage CD in this population |
| Levine et al., 2019 (91) | The study compared the efficacy and tolerability of the CDED coupled with PEN to EEN in children with mild to moderate CD. The combination of CDED and PEN was better tolerated and produced sustained remission in a significantly higher proportion of patients than EEN, with associated changes in the fecal microbiome. The study supports using CDED plus PEN to induce remission in children with CD | |
| Yanai et al., 2022 (50) | The CDED with or without partial enteral nutrition effectively induced and maintained remission in adults with mild-to-moderate CD, according to an open-label, pilot, randomized trial conducted in Israel. The study found that CDED may lead to endoscopic remission and should be further evaluated in a powered randomized controlled trial. No serious adverse events or treatment-related adverse events were reported in either group | |
| Diet (specific carbohydrate diet) | Suskind et al., 2014 (92) | The study reviewed the medical records of seven pediatric CD patients who followed the SCD without immunosuppressive medications. The results showed notable symptom resolution and improvements in laboratory indices, suggesting that SCD and other low-complex carbohydrate diets may be potential therapeutic options for pediatric CD. However, further prospective studies are needed to assess their safety and efficacy |
| Suskind et al., 2016 (93) | This study investigated the perceived benefits of SCD in patients with IBD. A survey was conducted on 80 participants with IBD who had been following the SCD for at least one month. The results showed that SCD was associated with improved abdominal pain, bloating, diarrhea, and stool consistency. The participants also reported an overall improvement in their quality of life, with 80% reporting a significant improvement. The study suggests that SCD may be an effective therapeutic option for patients with IBD, and further research is needed to evaluate its efficacy and safety | |
| Lewis et al., 2021 (94) | This study aimed to compare the effectiveness of SCD to the MD as a treatment for CD with mild to moderate symptoms. The study randomized 194 patients, and the results showed that the SCD was not superior to the MD in achieving symptomatic remission, fecal calprotectin response, and C-reactive protein response. Given the results, the MD may be preferred to the SCD for most patients with CD with mild to moderate symptoms | |
| Diet (anti-inflammatory diet) | Peter et al., 2020 (54) | The MELODY trial aims to investigate whether a dietary intervention during the last trimester of pregnancy can beneficially shift the microbiome of pregnant women with CD and their babies, thereby promoting a robust, effective immune system during a critical time of immune system development. The study hopes to create new opportunities to foster a healthy microbiome in the offspring at high risk of other immune-mediated diseases, potentially reducing their risk later in life |
| Olendzki et al., 2014 (95) | The study reports on a case series investigating the IBD-AID as a nutritional therapy for IBD. The IBD-AID involves restricting certain carbohydrates, consuming pre-and probiotic foods, and modifying dietary fatty acids. The case series provides preliminary evidence that the IBD-AID may be a beneficial adjunct therapy for IBD. Patients experienced an improvement in symptoms and inflammatory markers while following the diet | |
| Microbiome | Morgan et al., 2012 (96) | The study aimed to identify the dysfunctions of microbial metabolism in the gastrointestinal microbiome during IBD. The researchers analyzed the microbiota of intestinal biopsies and stool samples from 231 IBD and healthy subjects and followed up a subset using shotgun metagenomics. They found that the composition and abundance of specific bacterial taxa were altered in IBD patients compared to healthy individuals. Additionally, the researchers identified differences in microbial metabolic pathways associated with IBD, including alterations in amino acid and carbohydrate metabolism. These findings suggest that microbial dysfunction plays a significant role in the pathogenesis of IBD and could be a target for future therapies |
| Glassner et al., 2020 (97) | The study discusses the potential role of the microbiome in the development, progression, and treatment of IBD, a chronic immune-mediated disease affecting the gastrointestinal tract. The gut microbiome differs in patients with IBD compared to healthy control subjects. Evidence implicates microbiota-modulating risk factors such as antibiotic use, cigarette smoking, levels of sanitation, and diet in the pathogenesis of IBD. The article suggests that microbiome-modulating interventions such as probiotics, prebiotics, antibiotics, fecal microbiota transplantation, and gene manipulation may have potential benefits in treating IBD | |
| Underhill and Braun, 2022 (98) | The paper discusses recent advances in understanding the role of the fungal microbiome in IBD pathogenesis and disease activity. While the bacterial microbiome has been studied extensively, current tools have enabled the study of host genetics and metagenomics of host-fungal interaction. The article presents strong evidence linking specific fungal taxa, such as Candida and Malassezia, with cellular and molecular pathways of IBD disease biology. The study also suggests that some disease-participatory bacteria and fungi may act not directly on the host but via their fungal-bacterial ecologic interactions. The authors hope these insights will facilitate the inclusion of the fungal community in basic and translational IBD research | |
| Breastfeeding | Klement, 2004 (99) | Meta-analysis shows that breastfeeding is associated with lower risks of IBD, specifically CD, and UC. However, more high-quality research is needed to strengthen the validity of these findings |
| Barclay et al., 2009 (100) | This systematic review aimed to assess the evidence for the role of breastfeeding in the development of pediatric IBD. A total of 7 studies were included in the meta-analysis, which showed that breast milk exposure had a significant protective effect (OR, 0.69) in developing early-onset IBD. However, the quality of existing data could be better, and further well-designed prospective studies are needed to confirm these findings | |
| Lautenschlager et al., 2020 (101) | The study found that breastfeeding for less than 6 months was associated with a decreased risk for UC/IC. IBD patients reported fewer pet animals in the household than the control group, and the presence of cats or dogs and pet rodents before the age of 20 was inversely associated with the risk for UC/IC. The study highlights the importance of environmental factors in the pathogenesis of IBD. It suggests a protective effect of possessing pet animals in the household and short breastfeeding regarding the onset of UC/IC | |
| Smoking | Rubin and Hanauer, 2000 (40) | The paper discusses the well-established associations between smoking and IBD, where smoking is associated with CD and non-smoking with UC. It also highlights that smoking has a negative effect on the course of CD and may improve disease severity or have a protective effect in some patients with UC. However, the mechanism of smoking affects IBD is poorly understood, and studies have not adequately explained the observed clinical patterns. Although nicotine has been assumed to be the active agent in these associations, clinical trials of nicotine therapy have shown limited benefits and significant side effects. Topical delivery systems for nicotine therapy are currently under development and require further clinical trials |
| Mahid et al., 2006 (39) | The paper examines the relationship between smoking and IBD, specifically CD and UC. The study searched multiple healthcare databases from 1980 to 2006 and found evidence of an association between current smoking and CD, as well as former smoking and UC. The study also found that current smoking had a protective effect on the development of UC when compared to controls. This meta-analysis confirms that smoking is an essential environmental factor in IBD, with differing results in UC and CD. The study provides an estimate of the effect of smoking on both forms of IBD using predefined inclusion criteria and testing for homogeneity | |
| Jones et al., 2020 (102) | This study investigated whether smoking is a causal factor in developing IBD using observational techniques followed by Mendelian randomization. The results showed an association between smoking and IBD, with an increased risk of IBD in those who have ever smoked, an increased risk of CD, and decreased risk of UC in current smokers. However, the Mendelian randomization analyses found little evidence that smoking affects the development of IBD. Therefore, the overall results suggest that smoking does not causally influence the risk of IBD | |
| Physical activity | Gatt et al., 2019 (103) | The study aimed to determine the impact of IBD diagnosis on fitness levels and physical activity engagement among patients with IBD. One hundred fifty-eight patients diagnosed with IBD in the previous 18 months were recruited. The results showed that patients were significantly less physically active after an IBD diagnosis, which was more apparent in UC patients. Identifying risk factors associated with losing fitness levels would help address the reduced quality of life in IBD patients |
| Eckert et al., 2019 (104) | This paper is a scoping review that aims to collect data on the effects of structured physical activity interventions on validated clinical parameters of IBD and health-related symptoms and to develop actionable recommendations for IBD patients. The authors found that structured physical activity interventions can improve disease activity, quality of life, and fatigue in IBD patients. The article provides practical recommendations for healthcare providers to help IBD patients incorporate physical activity into their treatment plans | |
| van Erp et al., 2021 (105) | The study aimed to assess the effect of a personalized, intensive exercise program on fatigue, HRQoL, and cardiorespiratory fitness in patients with quiescent IBD and severe fatigue. The results showed that the personalized exercise program significantly improved fatigue and HRQoL in patients with quiescent IBD but not in cardiorespiratory fitness. The study suggests that a personalized exercise program can be an effective complementary therapy for improving fatigue and HRQoL in patients with quiescent IBD | |
| Stress/anxiety | Gracie et al., 2019 (106) | The brain-gut axis plays a role in developing and reporting gastrointestinal symptoms in functional gastrointestinal disorders, including IBS. In IBD, reporting IBS-type symptoms by patients with quiescent disease is common. It is associated with psychological disorders, impaired quality of life, and increased healthcare use. Data from observational studies suggest that psychological disorders might be related to relapse of disease activity in IBD and that inflammatory activity is associated with developing new psychological disorders. The article suggests that treatments targeting disordered brain-gut axis activity, including psychological therapies and antidepressants, might improve symptoms and quality of life in some patients with IBD. Further well-designed trials are needed to evaluate the benefit of these treatments |
| Sun et al., 2019 (107) | The paper discusses the impact of stress on IBD and how stress affects IBD, including impaired intestinal barrier function, disturbance of the gut microbiota, intestinal dysmotility, and immune and immune and neuroendocrine dysfunction. The authors emphasize the importance of mental healing and discuss the value of approaches targeting stress in clinical management to develop enhanced strategies for the prevention and treatment of IBD in both children and adults. Evidence from clinical and experimental studies suggests stress is a promoting or relapsing factor for IBD. Recent studies have reported increased anxiety or depression in children and adults with IBD | |
| Bisgaard et al., 2022 (108) | This paper reviews the literature on the co-occurrence of depression and anxiety in patients with IBD and the mechanisms underlying this association. The authors discuss the effect of psychological stress on the onset and course of IBD and outline possible tools such as changes in brain signaling, gut dysbiosis, and genetics. The review also examines the potential effects of treating depression and anxiety on the risk and course of IBD and the effects of IBD treatment on psychiatric comorbidity | |
| Pollution | Kaplan et al., 2010 (109) | This study investigated the association between ambient air pollution and the incidence of IBD, including CD and UC. The study used the UK THIN database and found that exposure to NO2 was associated with a higher risk of CD in individuals aged 23 years or younger. In comparison, SO2 exposure was associated with a higher risk of UC in individuals aged 25 years or younger. However, overall air pollution exposure was not related to the incidence of IBD. The study suggests that future studies should investigate the age-specific effects of air pollution exposure on IBD risk |
| Opstelten et al., 2016 (110) | The study aimed to investigate the association between exposure to ambient air pollution and the risk of IBD. The researchers conducted a nested case-control study in seven European countries, including 1,033 incident IBD cases and 1,733 matched controls. The study found no association between long-term exposure to ambient air pollution, including particulate matter (PM10, PM2.5) and NOx, and the risk of IBD. However, a weak association was observed between exposure to PM2.5 and the risk of CD, particularly among nonsmokers. The authors suggest that further research is needed to investigate the potential association between air pollution and IBD risk, particularly for CD | |
| Ding et al., 2022 (85) | This study evaluated the association between daily air pollution and IBD in Hefei, China. The study found that exposure to PM2.5, O3, and CO significantly increased the risk of IBD. Each increase of 10 mg/m3 in PM2.5/O3 and 0.1 mg/m3 in CO increased the risk of IBD. The most potent effects of these pollutants on IBD were observed in lag2-lag3, lag3, and lag2, respectively. The study suggests that air pollutants increase the risk of IBD patients in Hefei, China, and that warm seasons are the most susceptible to exposure. However, further studies are needed to verify the associations | |
| Antibiotics | Shaw et al., 2010 (111) | The paper reports a study to determine if early use of antibiotics was associated with developing IBD in childhood. The researchers conducted a nested case-control analysis of the University of Manitoba IBD Epidemiologic Database. They found that children diagnosed with IBD were more likely to have used antibiotics in their first year. Those who received one or more dispensations of antibiotics were at 2.9 times the odds of being an IBD case |
| Theochari et al., 2018 (112) | This systematic review aimed to assess the evidence suggesting an association between antibiotic exposure and the new onset of IBD. Fifteen observational studies, including 8,748 patients diagnosed with IBD, were analyzed. Antibiotic exposure was primarily associated with CD but not with UC, and penicillins, cephalosporins, metronidazole, and fluoroquinolones were commonly associated with the onset of CD. However, available data need to be granular enough to reach definitive conclusions, and clinicians should be cautious when prescribing certain antibiotics to patients with a strong family history of IBD | |
| Nguyen et al., 2020 (113) | The paper investigated the association between antibiotic therapy and IBD in a large population-based study in Sweden. The study found that higher cumulative exposure to systemic antibiotic therapy, particularly broad-spectrum antibiotics, may be associated with a greater risk of new-onset IBD and its subtypes. The study further emphasizes antibiotic stewardship to prevent dysbiosis-related chronic diseases, including IBD |
IBD, inflammatory bowel disease; CD, Crohn’s disease; FODMAPS, fermentable oligosaccharides, disaccharides, monosaccharides, and polyol; MD, Mediterranean diet; IBS, irritable bowel syndrome; CDED, CD exclusion diet; PEN, partial enteral nutrition; EEN, exclusive enteral nutrition; SCD, specific carbohydrate diet; AID, anti-inflammatory diet; UC, ulcerative colitis; OR, odds ratio; IC, indeterminate colitis; HRQoL, health-related quality of life; THIN, The Health Improvement Network; NO2, nitrogen dioxide; SO2, sulfur dioxide; NOx, nitrogen oxides.
Epigenome in IBDs
All these topics mentioned up to now are responsible for shaping the epigenome. Epigenetic programming begins at fertilization and continues throughout life (114). Food intake during pregnancy has been shown to affect epigenetic reprogramming steps during the early development of offspring, with effects lasting up to 2 generations (115). Indeed, as we have seen, research advances reinforce the idea that epimutations (disease-associated epigenetic changes) are induced by multiple environmental factors (116). Some factors relevant to IBD are smoking habits and diet, which play a vital role in the disease, both by direct action and for the modulation of the intestinal microbiome, and also have epigenetic activity (4).
In addition, there is evidence that various environmental factors contribute to age-acquired epigenetic changes (116). Studies of identical twins show that by age 65, almost 70% of the immune epigenome is environmental (117). Hence epigenetic factors could mediate gene-environment interactions involved in resistant output and disease susceptibility, which makes epigenetics a key mechanism in IBD pathogenesis and other chronic and immune-mediated diseases (115).
The epigenome can be considered stable because epigenetic marks are replicated during mitosis. However, random and environmental factors can cause dynamic changes to the epigenome over time (118). A study of epigenetics examines how environment and behavior influence gene activity, resulting in phenotypic differences (118). In 1942, Waddington introduced the epigenetic concept (119). Epigenetic mechanisms include DNA methylation, histone modification, ncRNAs, and nucleosome positioning (12). Among these mechanisms, ncRNAs are among the most extensively studied in IBD research (12). Natural epigenetic modifications play a vital role in organism function, but if they occur incorrectly, they can cause serious health consequences (116).
DNA modifications: DNA methylation and histone modifications
Epigenetic changes modify gene expression without affecting the DNA sequence (120). Chromatin, a complex of DNA and histone proteins, plays an epigenetic role in transcriptional regulation (121). Several protein complexes that can remodel the chromatin architecture are involved in regulating chromatin by modifying histones or DNA (121). Post-transcriptional modifications to histones allow heterochromatin to be tightly packed or more accessible (122). Nucleic acids form chromatin: genomic DNA, different RNA types, and histone proteins, such as H2A, H2B, H3, H4, and H1, besides other molecules (123).
DNA methylation is stably transferred through repetitive cell divisions, leading to the capacity to permanently transmit epigenetic information during an individual’s lifetime (121,124,125). It also involves adding a methyl group onto the cytosine’s 5’ position to form 5-methylcytosine, one of the most studied epigenetic mechanisms (125). This occurs at a CpG site, where a cytosine nucleotide precedes a guanine nucleotide and is catalyzed by DNA methyltransferases (126). The CpG island is defined as DNA regions that are ≥200 bp long and show a CG:GC ratio ≥0.6. DNA methylation typically prevents transcription factors from producing gene promoters, resulting in gene silencing (4,126). Interestingly, dietary substrates and nutrients drive DNA methylation by providing cofactors (4,126). Vitamins B12 and folate, for instance, act as cofactors, contributing to DNA methylation (37,114). Compared to healthy subjects, IBD patients acquire DNA methylation changes at the cell, tissue, and mostly immune level (37). Moreover, the profile differs between UC and CD patients (4).
In primary human epithelial cells obtained from mucosal biopsies of pediatric IBD patients, genome study analysis revealed differences in DNA methylation and transcriptional changes compared with control samples (126). Pediatric IBD epithelium showed significant overlap with those undergoing methylation changes during fetal intestine development, including key genes in intestinal epithelium related to immunity (114). Ventham et al. [2016] found 439 differentially methylated positions and five differentially methylated regions, mainly vacuole-membrane protein 1 (VMP1) (118). The major differentially methylated position was RPS6KA2, a ribosomal kinase in the serine/threonine kinase family that regulates cell growth and proliferation (118). In peripheral blood mononuclear cells, the promoter region of TRIM39-RPP2 was also significantly hypomethylated in colonic mucosa from pediatric UC patients (127). On the other hand, TRAF6 hypermethylation was observed. Methylation patterns observed in blood samples from the CD change the patterns during disease development/progression or in acute inflammation (128). In addition, the increased risk of colorectal cancer in IBD patients was associated with global hypermethylation, probably driven by inflammation (129).
The importance of correlating blood methylation findings with microbiome assessment results is worth mentioning due to the dysbiosis in patients with IBD. A large multi-omic study evaluated the microbiome of patients with CD and UC for one year and found several species of bacteria not yet cataloged, in addition to a reduction in enzymes, specific fatty acids, and some vitamins such as B5 and B3 (130). More studies like this are needed better understand the role of epigenome associated with the patient’s microbiome.
Proteins called histones are comparatively large and basic proteins that play an important role in the nucleosome of every cell (122,123). Nucleosomes constitute the functional and structural units of chromatin. Due to the nucleosome’s structural characteristics, histone proteins can undergo post-translational modifications at their N-terminal tails, such as acetylation, methylation, phosphorylation, ubiquitination, propionylation, butyrylation, crotonylation, and sumoylation, among others (123,126). Over 400 chromatin-modifying enzymes are regulated by the modifications. Post-translational modifications on histones or DNA are catalyzed by epigenetic “writers”, such as DNA and histone methyltransferases and histone acetyltransferases. In contrast, dynamic modifications are removed by “erasers”, such as demethylases, histone deacetylases (HDACs), and ten-eleven translocation (TET) proteins (121). Experimental colitis in mice with dextran sulfate sodium (DSS) that have HDAC 1 and 2 depletion specific in intestinal epithelial increases the severity of colitis (122). Butyrate can reduce gut inflammation by acting as an HDAC inhibitor and by suppressing nuclear factor kappa B (NF-kB) activation (122). Additionally, Lactobacillus and Bifidobacterium can also affect DNA methylation by regulating methyl-donor availability through their folate production (122). Patients with IBD have lower levels of the catalytic histone methyltransferase subunit of polycomb repressive complex 2 (PRC2) and the enhancer of zeste homolog 2 (EZH2) (122). However, after DSS treatments, mice intestines were protected from inflammation by overexpressing EZH2, indicating the significance of polycomb complexes in intestinal regeneration following epithelial damage (122).
IBD has numerous complex causes, including environmental triggers and genetic predisposition. DNA modifications mentioned above demonstrate the critical role of studying these alterations in IBD models (in vivo) and patient samples. Interactions between the microbiota and the host most likely cause IBD (123). It is essential to comprehend the extent to which these defective responses are supported by epigenetic pathways. In mice and children with newly-diagnosed IBD, changes in the histone H3K4me3 level in intestinal cells revealed an epigenetic profile of their disease (123). The presence of commensal gut bacteria could prop these changes (131). This study demonstrates how epigenetic analyses can complement other methods for identifying epithelial abnormalities, despite the small number of patient samples. It also sheds new light on pathways by which microbiota might be predisposed to intestinal inflammation (123,131).
ncRNAs and miRNAs
ncRNAs are RNAs that are not translated into proteins but are correlated with cellular processes such as translation, RNA splicing, and gene and chromosome structure modulation (12). These molecules include small interfering RNA (siRNA), microRNA (miRNA), long non-coding RNA (lncRNA), and others. The most studied class of small ncRNA is the miRNAs (132). MiRNAs are a class of regulatory RNAs that suppress gene expression. They are single-stranded, noncoding RNA molecules that range in size from 19 to 24 nucleotides and are evolutionarily conserved (133).
Over 60% of human protein-coding genes are regulated by miRNAs (134). MiRNAs and ncRNAs are assumed to play a significant role in regulating metabolic pathways that influence disease progression in IBD (133). This may be seen at both transcriptional and post-transcriptional levels. Fisher and Lin [2016] compiled extensive literature showing miRNAs dysregulated in peripheral blood and mucosal tissue of IBD patients with activity disease (133). MiRNAs are all involved with disease onset and progression, regulating, for example, the Th17 and IL23 pathways and autophagy (12).
McKenna et al. [2010] showed in an animal model study that intestinal miRNAs regulate gut homeostasis (135). Dicer, a miRNA-processing enzyme, plays a vital role in the differentiation and function of the intestinal epithelium. In addition, Dicer-deficient mice have disorganized intestinal epithelial crypts with rapid jejunal epithelial migration, increased goblet cells, accelerated apoptosis, increased inflammation, and reduced epithelial barrier function with neutrophil and lymphocyte migration (135).
Another miRNA directly enrolled in IBD is miR-21. Recent studies have frequently shown the presence of this upregulated miRNA in both UC and CD (133). In peripheral blood and mucosal tissue samples from CD patients, miR-21 was overexpressed (136). Another study showed that mice with a genetic deletion of miR-21 were resistant to DSS induced colitis (137). However, the mechanisms of action of this miRNA in the IBD are still unknown (137).
Fisher and Lin [2016] state that miR-21 has at least three distinct routes that might have harmful consequences in IBD (133): (I) miR-21 may enhance permeability in response to epithelial injury; (II) miR-21 may defend against free radical-induced apoptosis; and (III) miR-21 has been linked to irreversible fibrosis in IBD; specific disease models have indicated that fibrosis is dependent on elevated levels of TNF and subsequent elevation of this miRNA.
Another miRNA involved in essential changes in intestinal epithelial cells (IECs) is miR-200b. This miRNA was significantly decreased in inflamed mucosa of IBD when compared with their adjacent normal tissue, especially for UC (138). Chen et al. [2013] identified that the downregulation of miR-200b expression was correlated with upregulation genes that act on the process of epithelial-mesenchymal transition (EMT), promoting a loss of IECs (138). There is increasing evidence that one of the primary factors in the etiology of IBD is the loss of IECs. Inhibiting EMT and encouraging IEC proliferation, miR-200b is crucial for preserving intestinal epithelium’s integrity (138).
The immune system’s control is another instance of how miRNAs have an epigenetic effect. The interferon gamma locus must be epigenetically silenced for Th2 cells to differentiate (138). Additionally, Th1 and Th17 cell-mediated immune responses are regulated by clusters of up-regulating miRNAs by CD-associated variants of the NOD2 gene expressed by dendritic cells (115).
Another critical and complicating factor in the epigenetic regulation of miRNAs is their involvement in many physiological processes, such as differentiation, proliferation, apoptosis, and the development of other diseases. Some miRNAs and their regulation have been reported in other pathological disorders, including muscular dystrophy, diabetes, and various cancers (132). For example, miR-27a expression was increased in the peripheral blood of CD and UC (139) and breast cancer cell lines and colon cancer cells (139).
Del Follo-Martinez et al. [2013] evaluated the action of a combination of resveratrol and quercetin, derived from apples, grapes, and onions, on colon cancer cells (139). They observed a reduction in the miR-27a levels, which leads to ZBTB10 gene expression and apoptosis induction in HT-29 colon cancer cells. Certain natural compounds, such as phenolic and terpenoid compounds (e.g., curcumin, boswellic acid), can modulate inflammatory pathways by regulating miRNAs involved in processes such as proliferation, differentiation, cell death, angiogenesis, and metastasis formation (140). In this context, these compounds may also be beneficial for treating IBD (140).
On the other hand, the major obstacle in using miRNAs for diagnosis and the therapeutic target is their low specificity, considering that the same miRNA can be modulated in several diseases (132). Moreover, identifying the targets of these miRNAs will provide additional insight into the pathogenesis of IBD (133).
Increasing our understanding of interactions’ pathways among genetic, environmental, and microbial factors can answer why only some people with a specific genetic risk develop IBD (29).
Conclusions
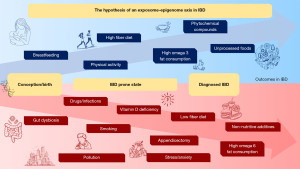
This review focused on some essential aspects of IBD pathogenesis. While looking at this multifactorial disease in context, we noticed the role of “omics” changes in IBD. Genomics could not explain the illness by itself. This allowed the investigation of other pathways, such as the epigenome, microbiome, diet, and exposome, to achieve the best outcome for IBD patients. Different elements of the exposome, such as the maternal lifestyle, microbiota, diet, smoking, physical activity, infection, vitamin D, antibiotics, anti-inflammatory drugs, and different drugs, may induce epigenetic changes related to IBD. Psychological stress/anxiety/depression, oral contraception, geographical location, and other factors potentially predicted the disease. The environmental factors, such as the exposome, still need to be clarified as to how their duration, timing, and frequency may be able to modulate IBD outcomes (13,18,141). Finally, the exposome may become a tool to predict flares and contribute to developing electronic technologies based on artificial intelligence, algorithms, and machine learning. These mathematical tools will make possible to record patients’ exposure to environmental factors continuously and to better understand these complex immune-mediated diseases. In conclusion, the hypothesis of an exposome-diet-epigenome axis (Figure 1) combines the main findings described so far that can influence and lead to IBD outcomes, from diagnosis to degree of severity.
Acknowledgments
We thank Dr. Tristan Torriani [Assistant Professor of School of Applied Sciences from the University of Campinas (Unicamp), Limeira, São Paulo, Brazil] for revising the English version of our manuscript.
Funding: This work was supported by the National Council for Scientific and Technological Development (CNPq) (grant number #302557/2021-0 for R.F.L., grant number #140520/2019-8 for K.M.S.), the São Paulo Research Foundation (FAPESP) (grant number #2018/17832-2 for P.L.R.M.), and the Unicamp Development Foundation (Grant number #519.292 for B.A.G.R.).
Footnote
Provenance and Peer Review: This article was commissioned by the editorial office, Digestive Medicine Research for the series “Evidence of Epigenetics in Inflammatory Bowel Diseases”. The article has undergone external peer review.
Reporting Checklist: The authors have completed the Narrative Review reporting checklist. Available at https://dmr.amegroups.com/article/view/10.21037/dmr-22-85/rc
Peer Review File: Available at https://dmr.amegroups.com/article/view/10.21037/dmr-22-85/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dmr.amegroups.com/article/view/10.21037/dmr-22-85/coif). The series “Evidence of Epigenetics in Inflammatory Bowel Diseases” was commissioned by the editorial office without any funding or sponsorship. R.F.L. serves as an unpaid editorial board member of Digestive Medicine Research from October 2021 to September 2023. R.F.L. served as the unpaid Guest Editor of the series. B.A.G.R. receives a postdoctoral scholarship from the Unicamp Development Foundation (grant number #519.292). K.M.S. receives a doctoral scholarship from the National Council for Scientific and Technological Development (CNPq) (grant number #140520/2019-8). P.L.R.M. receives a postdoctoral scholarship from the São Paulo Research Foundation (FAPESP) (grant number #2018/17832-2). R.F.L. receives support from the National Council for Scientific and Technological Development (CNPq) (grant number #302557/2021-0). The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Marafini I, Monteleone G. Precision Medicine in Inflammatory Bowel Diseases. Front Pharmacol 2021;12:653924. [Crossref] [PubMed]
- Lavelle A, Sokol H. Gut microbiota-derived metabolites as key actors in inflammatory bowel disease. Nat Rev Gastroenterol Hepatol 2020;17:223-37. [Crossref] [PubMed]
- de Souza HS, Fiocchi C. Immunopathogenesis of IBD: current state of the art. Nat Rev Gastroenterol Hepatol 2016;13:13-27. [Crossref] [PubMed]
- Vieujean S, Caron B, Haghnejad V, et al. Impact of the Exposome on the Epigenome in Inflammatory Bowel Disease Patients and Animal Models. Int J Mol Sci 2022;23:7611. [Crossref] [PubMed]
- Ng SC, Shi HY, Hamidi N, et al. Worldwide incidence and prevalence of inflammatory bowel disease in the 21st century: a systematic review of population-based studies. Lancet 2017;390:2769-78. [Crossref] [PubMed]
- Nurmi E, Haapamäki J, Paavilainen E, et al. The burden of inflammatory bowel disease on health care utilization and quality of life. Scand J Gastroenterol 2013;48:51-7. [Crossref] [PubMed]
- Gibson PR, Weston AR, Shann A, et al. Relationship between disease severity, quality of life and health-care resource use in a cross-section of Australian patients with Crohn's disease. J Gastroenterol Hepatol 2007;22:1306-12. [Crossref] [PubMed]
- Silverberg MS, Satsangi J, Ahmad T, et al. Toward an integrated clinical, molecular and serological classification of inflammatory bowel disease: report of a Working Party of the 2005 Montreal World Congress of Gastroenterology. Can J Gastroenterol 2005;19 Suppl A:5A-36A.
- Fofanova TY, Petrosino JF, Kellermayer R. Microbiome-Epigenome Interactions and the Environmental Origins of Inflammatory Bowel Diseases. J Pediatr Gastroenterol Nutr 2016;62:208-19. [Crossref] [PubMed]
- Amatullah H, Jeffrey KL. Epigenome-metabolome-microbiome axis in health and IBD. Curr Opin Microbiol 2020;56:97-108. [Crossref] [PubMed]
- Annese V. Genetics and epigenetics of IBD. Pharmacol Res 2020;159:104892. [Crossref] [PubMed]
- Zeng Z, Mukherjee A, Zhang H. From Genetics to Epigenetics, Roles of Epigenetics in Inflammatory Bowel Disease. Front Genet 2019;10:1017. [Crossref] [PubMed]
- Cleynen I, Vermeire S. The genetic architecture of inflammatory bowel disease: past, present and future. Curr Opin Gastroenterol 2015;31:456-63. [Crossref] [PubMed]
- Loddo I, Romano C. Inflammatory Bowel Disease: Genetics, Epigenetics, and Pathogenesis. Front Immunol 2015;6:551. [Crossref] [PubMed]
- Liu JZ, van Sommeren S, Huang H, et al. Association analyses identify 38 susceptibility loci for inflammatory bowel disease and highlight shared genetic risk across populations. Nat Genet 2015;47:979-86. [Crossref] [PubMed]
- de Lange KM, Moutsianas L, Lee JC, et al. Genome-wide association study implicates immune activation of multiple integrin genes in inflammatory bowel disease. Nat Genet 2017;49:256-61. [Crossref] [PubMed]
- Schirmer M, Garner A, Vlamakis H, et al. Microbial genes and pathways in inflammatory bowel disease. Nat Rev Microbiol 2019;17:497-511. [Crossref] [PubMed]
- Graham DB, Xavier RJ. Pathway paradigms revealed from the genetics of inflammatory bowel disease. Nature 2020;578:527-39. [Crossref] [PubMed]
- Ogura Y, Bonen DK, Inohara N, et al. A frameshift mutation in NOD2 associated with susceptibility to Crohn's disease. Nature 2001;411:603-6. [Crossref] [PubMed]
- Hugot JP. CARD15/NOD2 mutations in Crohn’s disease. Ann N Y Acad Sci 2006;1072:9-18. [Crossref] [PubMed]
- Uhlig HH, Muise AM. Clinical Genomics in Inflammatory Bowel Disease. Trends Genet 2017;33:629-41. [Crossref] [PubMed]
- Franke A, McGovern DP, Barrett JC, et al. Genome-wide meta-analysis increases to 71 the number of confirmed Crohn's disease susceptibility loci. Nat Genet 2010;42:1118-25. [Crossref] [PubMed]
- McGovern DP, Gardet A, Törkvist L, et al. Genome-wide association identifies multiple ulcerative colitis susceptibility loci. Nat Genet 2010;42:332-7. [Crossref] [PubMed]
- Festen EA, Goyette P, Green T, et al. A meta-analysis of genome-wide association scans identifies IL18RAP, PTPN2, TAGAP, and PUS10 as shared risk loci for Crohn's disease and celiac disease. PLoS Genet 2011;7:e1001283. [Crossref] [PubMed]
- González-Serna D, Ochoa E, López-Isac E, et al. A cross-disease meta-GWAS identifies four new susceptibility loci shared between systemic sclerosis and Crohn's disease. Sci Rep 2020;10:1862. [Crossref] [PubMed]
- Jostins L, Ripke S, Weersma RK, et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature 2012;491:119-24. [Crossref] [PubMed]
- Hornschuh M, Wirthgen E, Wolfien M, et al. The role of epigenetic modifications for the pathogenesis of Crohn's disease. Clin Epigenetics 2021;13:108. [Crossref] [PubMed]
- Zuckerman MK, Harper KN, Barrett R, et al. The evolution of disease: anthropological perspectives on epidemiologic transitions. Glob Health Action 2014;7:23303. [Crossref] [PubMed]
- Kaplan GG, Ng SC. Understanding and Preventing the Global Increase of Inflammatory Bowel Disease. Gastroenterology 2017;152:313-321.e2. [Crossref] [PubMed]
- Kaplan GG, Windsor JW. The four epidemiological stages in the global evolution of inflammatory bowel disease. Nat Rev Gastroenterol Hepatol 2021;18:56-66. [Crossref] [PubMed]
- Yan Z, Liu Y, Zhang T, et al. Analysis of Microplastics in Human Feces Reveals a Correlation between Fecal Microplastics and Inflammatory Bowel Disease Status. Environ Sci Technol 2022;56:414-21. [Crossref] [PubMed]
- Vedamurthy A, Ananthakrishnan AN. Influence of Environmental Factors in the Development and Outcomes of Inflammatory Bowel Disease. Gastroenterol Hepatol (N Y) 2019;15:72-82. [PubMed]
- Ramos GP, Papadakis KA. Mechanisms of Disease: Inflammatory Bowel Diseases. Mayo Clin Proc 2019;94:155-65. [Crossref] [PubMed]
- Carbonnel F, Jantchou P, Monnet E, et al. Environmental risk factors in Crohn's disease and ulcerative colitis: an update. Gastroenterol Clin Biol 2009;33:S145-S157. [Crossref] [PubMed]
- Yan J, Wang L, Gu Y, et al. Dietary Patterns and Gut Microbiota Changes in Inflammatory Bowel Disease: Current Insights and Future Challenges. Nutrients 2022;14:4003. [Crossref] [PubMed]
- Li T, Qiu Y, Yang HS, et al. Systematic review and meta-analysis: Association of a pre-illness Western dietary pattern with the risk of developing inflammatory bowel disease. J Dig Dis 2020;21:362-71. [Crossref] [PubMed]
- Aleksandrova K, Romero-Mosquera B, Hernandez V. Diet, Gut Microbiome and Epigenetics: Emerging Links with Inflammatory Bowel Diseases and Prospects for Management and Prevention. Nutrients 2017;9:962. [Crossref] [PubMed]
- Lunney PC, Leong RW. Review article: Ulcerative colitis, smoking and nicotine therapy. Aliment Pharmacol Ther 2012;36:997-1008. [Crossref] [PubMed]
- Mahid SS, Minor KS, Soto RE, et al. Smoking and inflammatory bowel disease: a meta-analysis. Mayo Clin Proc 2006;81:1462-71. [Crossref] [PubMed]
- Rubin DT, Hanauer SB. Smoking and inflammatory bowel disease. Eur J Gastroenterol Hepatol 2000;12:855-62. [Crossref] [PubMed]
- Berkowitz L, Schultz BM, Salazar GA, et al. Impact of Cigarette Smoking on the Gastrointestinal Tract Inflammation: Opposing Effects in Crohn's Disease and Ulcerative Colitis. Front Immunol 2018;9:74. [Crossref] [PubMed]
- Pekow JR, Kwon JH. MicroRNAs in inflammatory bowel disease. Inflamm Bowel Dis 2012;18:187-93. [Crossref] [PubMed]
- Nie L, Wu HJ, Hsu JM, et al. Long non-coding RNAs: versatile master regulators of gene expression and crucial players in cancer. Am J Transl Res 2012;4:127-50. [PubMed]
- de Castro MM, Pascoal LB, Steigleder KM, et al. Role of diet and nutrition in inflammatory bowel disease. World J Exp Med 2021;11:1-16. [Crossref] [PubMed]
- Adolph TE, Zhang J. Diet fuelling inflammatory bowel diseases: preclinical and clinical concepts. Gut 2022;71:2574-86. [Crossref] [PubMed]
- Astore C, Nagpal S, Gibson G. Mendelian Randomization Indicates a Causal Role for Omega-3 Fatty Acids in Inflammatory Bowel Disease. Int J Mol Sci 2022;23:14380. [Crossref] [PubMed]
- Di'Narzo AF, Houten SM, Kosoy R, et al. Integrative Analysis of the Inflammatory Bowel Disease Serum Metabolome Improves Our Understanding of Genetic Etiology and Points to Novel Putative Therapeutic Targets. Gastroenterology 2022;162:828-843.e11. [Crossref] [PubMed]
- Cholewski M, Tomczykowa M, Tomczyk M. A Comprehensive Review of Chemistry, Sources and Bioavailability of Omega-3 Fatty Acids. Nutrients 2018;10:1662. [Crossref] [PubMed]
- Connors J, Basseri S, Grant A, et al. Exclusive Enteral Nutrition Therapy in Paediatric Crohn's Disease Results in Long-term Avoidance of Corticosteroids: Results of a Propensity-score Matched Cohort Analysis. J Crohns Colitis 2017;11:1063-70. [Crossref] [PubMed]
- Yanai H, Levine A, Hirsch A, et al. The Crohn's disease exclusion diet for induction and maintenance of remission in adults with mild-to-moderate Crohn's disease (CDED-AD): an open-label, pilot, randomised trial. Lancet Gastroenterol Hepatol 2022;7:49-59. [Crossref] [PubMed]
- Sigall-Boneh R, Pfeffer-Gik T, Segal I, et al. Partial enteral nutrition with a Crohn's disease exclusion diet is effective for induction of remission in children and young adults with Crohn's disease. Inflamm Bowel Dis 2014;20:1353-60. [Crossref] [PubMed]
- Bischoff SC, Bager P, Escher J, et al. ESPEN guideline on Clinical Nutrition in inflammatory bowel disease. Clin Nutr 2023;42:352-79. [Crossref] [PubMed]
- Cox SR, Lindsay JO, Fromentin S, et al. Effects of Low FODMAP Diet on Symptoms, Fecal Microbiome, and Markers of Inflammation in Patients With Quiescent Inflammatory Bowel Disease in a Randomized Trial. Gastroenterology 2020;158:176-188.e7. [Crossref] [PubMed]
- Peter I, Maldonado-Contreras A, Eisele C, et al. A dietary intervention to improve the microbiome composition of pregnant women with Crohn's disease and their offspring: The MELODY (Modulating Early Life Microbiome through Dietary Intervention in Pregnancy) trial design. Contemp Clin Trials Commun 2020;18:100573. [Crossref] [PubMed]
- Sender R, Fuchs S, Milo R. Revised Estimates for the Number of Human and Bacteria Cells in the Body. PLoS Biol 2016;14:e1002533. [Crossref] [PubMed]
- Zuo T, Ng SC. The Gut Microbiota in the Pathogenesis and Therapeutics of Inflammatory Bowel Disease. Front Microbiol 2018;9:2247. [Crossref] [PubMed]
- Tamburini S, Shen N, Wu HC, et al. The microbiome in early life: implications for health outcomes. Nat Med 2016;22:713-22. [Crossref] [PubMed]
- Casén C, Vebø HC, Sekelja M, et al. Deviations in human gut microbiota: a novel diagnostic test for determining dysbiosis in patients with IBS or IBD. Aliment Pharmacol Ther 2015;42:71-83. [Crossref] [PubMed]
- O'Hara AM, Shanahan F. The gut flora as a forgotten organ. EMBO Rep 2006;7:688-93. [Crossref] [PubMed]
- Kelly CJ, Zheng L, Campbell EL, et al. Crosstalk between Microbiota-Derived Short-Chain Fatty Acids and Intestinal Epithelial HIF Augments Tissue Barrier Function. Cell Host Microbe 2015;17:662-71. [Crossref] [PubMed]
- Blaak EE, Canfora EE, Theis S, et al. Short chain fatty acids in human gut and metabolic health. Benef Microbes 2020;11:411-55. [Crossref] [PubMed]
- Parada Venegas D, De la Fuente MK, Landskron G, et al. Short Chain Fatty Acids (SCFAs)-Mediated Gut Epithelial and Immune Regulation and Its Relevance for Inflammatory Bowel Diseases. Front Immunol 2019;10:277. [Crossref] [PubMed]
- Derrien M, Vaughan EE, Plugge CM, et al. Akkermansia muciniphila gen. nov., sp. nov., a human intestinal mucin-degrading bacterium. Int J Syst Evol Microbiol 2004;54:1469-76. [Crossref] [PubMed]
- Lane ER, Zisman TL, Suskind DL. The microbiota in inflammatory bowel disease: current and therapeutic insights. J Inflamm Res 2017;10:63-73. [Crossref] [PubMed]
- Rizzello F, Spisni E, Giovanardi E, et al. Implications of the Westernized Diet in the Onset and Progression of IBD. Nutrients 2019;11:1033. [Crossref] [PubMed]
- Fan Y, Pedersen O. Gut microbiota in human metabolic health and disease. Nat Rev Microbiol 2021;19:55-71. [Crossref] [PubMed]
- Matsuoka K, Kanai T. The gut microbiota and inflammatory bowel disease. Semin Immunopathol 2015;37:47-55. [Crossref] [PubMed]
- Shaw KA, Bertha M, Hofmekler T, et al. Dysbiosis, inflammation, and response to treatment: a longitudinal study of pediatric subjects with newly diagnosed inflammatory bowel disease. Genome Med 2016;8:75. [Crossref] [PubMed]
- Mentella MC, Scaldaferri F, Pizzoferrato M, et al. Nutrition, IBD and Gut Microbiota: A Review. Nutrients 2020;12:944. [Crossref] [PubMed]
- Sokol H, Pigneur B, Watterlot L, et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc Natl Acad Sci U S A 2008;105:16731-6. [Crossref] [PubMed]
- Gevers D, Kugathasan S, Denson LA, et al. The treatment-naive microbiome in new-onset Crohn's disease. Cell Host Microbe 2014;15:382-92. [Crossref] [PubMed]
- Andoh A, Nishida A. Alteration of the Gut Microbiome in Inflammatory Bowel Disease. Digestion 2023;104:16-23. [Crossref] [PubMed]
- Rohr MW, Narasimhulu CA, Rudeski-Rohr TA, et al. Negative Effects of a High-Fat Diet on Intestinal Permeability: A Review. Adv Nutr 2020;11:77-91. [Crossref] [PubMed]
- Simopoulos AP. Omega-3 fatty acids in inflammation and autoimmune diseases. J Am Coll Nutr 2002;21:495-505. [Crossref] [PubMed]
- Devkota S, Wang Y, Musch MW, et al. Dietary-fat-induced taurocholic acid promotes pathobiont expansion and colitis in Il10-/- mice. Nature 2012;487:104-8. [Crossref] [PubMed]
- Lam YY, Ha CW, Hoffmann JM, et al. Effects of dietary fat profile on gut permeability and microbiota and their relationships with metabolic changes in mice. Obesity (Silver Spring) 2015;23:1429-39. [Crossref] [PubMed]
- Gearry RB, Irving PM, Barrett JS, et al. Reduction of dietary poorly absorbed short-chain carbohydrates (FODMAPs) improves abdominal symptoms in patients with inflammatory bowel disease-a pilot study. J Crohns Colitis 2009;3:8-14. [Crossref] [PubMed]
- Reddel S, Putignani L, Del Chierico F. The Impact of Low-FODMAPs, Gluten-Free, and Ketogenic Diets on Gut Microbiota Modulation in Pathological Conditions. Nutrients 2019;11:373. [Crossref] [PubMed]
- Godala M, Gaszyńska E, Zatorski H, et al. Dietary Interventions in Inflammatory Bowel Disease. Nutrients 2022;14:4261. [Crossref] [PubMed]
- Wang QP, Browman D, Herzog H, et al. Non-nutritive sweeteners possess a bacteriostatic effect and alter gut microbiota in mice. PLoS One 2018;13:e0199080. [Crossref] [PubMed]
- Chassaing B, Van de Wiele T, De Bodt J, et al. Dietary emulsifiers directly alter human microbiota composition and gene expression ex vivo potentiating intestinal inflammation. Gut 2017;66:1414-27. [Crossref] [PubMed]
- Laudisi F, Di Fusco D, Dinallo V, et al. The Food Additive Maltodextrin Promotes Endoplasmic Reticulum Stress-Driven Mucus Depletion and Exacerbates Intestinal Inflammation. Cell Mol Gastroenterol Hepatol 2019;7:457-73. [Crossref] [PubMed]
- Baer DJ, Stote KS, Henderson T, et al. The metabolizable energy of dietary resistant maltodextrin is variable and alters fecal microbiota composition in adult men. J Nutr 2014;144:1023-9. [Crossref] [PubMed]
- Liu Q, Shao W, Zhang C, et al. Organochloride pesticides modulated gut microbiota and influenced bile acid metabolism in mice. Environ Pollut 2017;226:268-76. [Crossref] [PubMed]
- Ding S, Sun S, Ding R, et al. Association between exposure to air pollutants and the risk of inflammatory bowel diseases visits. Environ Sci Pollut Res Int 2022;29:17645-54. [Crossref] [PubMed]
- Vasseur P, Dugelay E, Benamouzig R, et al. Dietary Patterns, Ultra-processed Food, and the Risk of Inflammatory Bowel Diseases in the NutriNet-Santé Cohort. Inflamm Bowel Dis 2021;27:65-73. [Crossref] [PubMed]
- Green N, Miller T, Suskind D, et al. A Review of Dietary Therapy for IBD and a Vision for the Future. Nutrients 2019;11:947. [Crossref] [PubMed]
- Gibson PR. Use of the low-FODMAP diet in inflammatory bowel disease. J Gastroenterol Hepatol 2017;32:40-2. [Crossref] [PubMed]
- Zhan YL, Zhan YA, Dai SX. Is a low FODMAP diet beneficial for patients with inflammatory bowel disease? A meta-analysis and systematic review. Clin Nutr 2018;37:123-9. [Crossref] [PubMed]
- Simões CD, Maganinho M, Sousa AS. FODMAPs, inflammatory bowel disease and gut microbiota: updated overview on the current evidence. Eur J Nutr 2022;61:1187-98. [Crossref] [PubMed]
- Levine A, Wine E, Assa A, et al. Crohn's Disease Exclusion Diet Plus Partial Enteral Nutrition Induces Sustained Remission in a Randomized Controlled Trial. Gastroenterology 2019;157:440-450.e8. [Crossref] [PubMed]
- Suskind DL, Wahbeh G, Gregory N, et al. Nutritional therapy in pediatric Crohn disease: the specific carbohydrate diet. J Pediatr Gastroenterol Nutr 2014;58:87-91. [Crossref] [PubMed]
- Suskind DL, Wahbeh G, Cohen SA, et al. Patients Perceive Clinical Benefit with the Specific Carbohydrate Diet for Inflammatory Bowel Disease. Dig Dis Sci 2016;61:3255-60. [Crossref] [PubMed]
- Lewis JD, Sandler RS, Brotherton C, et al. A Randomized Trial Comparing the Specific Carbohydrate Diet to a Mediterranean Diet in Adults With Crohn's Disease. Gastroenterology 2021;161:837-852.e9. Erratum in: Gastroenterology 2022;163:1473. [Crossref] [PubMed]
- Olendzki BC, Silverstein TD, Persuitte GM, et al. An anti-inflammatory diet as treatment for inflammatory bowel disease: a case series report. Nutr J 2014;13:5. [Crossref] [PubMed]
- Morgan XC, Tickle TL, Sokol H, et al. Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol 2012;13:R79. [Crossref] [PubMed]
- Glassner KL, Abraham BP, Quigley EMM. The microbiome and inflammatory bowel disease. J Allergy Clin Immunol 2020;145:16-27. [Crossref] [PubMed]
- Underhill DM, Braun J. Fungal microbiome in inflammatory bowel disease: a critical assessment. J Clin Invest 2022;132:e155786. [Crossref] [PubMed]
- Klement E, Cohen RV, Boxman J, et al. Breastfeeding and risk of inflammatory bowel disease: a systematic review with meta-analysis. Am J Clin Nutr 2004;80:1342-52. [Crossref] [PubMed]
- Barclay AR, Russell RK, Wilson ML, et al. Systematic review: the role of breastfeeding in the development of pediatric inflammatory bowel disease. J Pediatr 2009;155:421-6. [Crossref] [PubMed]
- Lautenschlager SA, Fournier N, Biedermann L, et al. The Influence of Breastfeeding, Cesarean Section, Pet Animals, and Urbanization on the Development of Inflammatory Bowel Disease: Data from the Swiss IBD Cohort Study. Inflamm Intest Dis 2020;5:170-9. [Crossref] [PubMed]
- Jones DP, Richardson TG, Davey Smith G, et al. Exploring the Effects of Cigarette Smoking on Inflammatory Bowel Disease Using Mendelian Randomization. Crohns Colitis 360 2020;2:otaa018. [Crossref] [PubMed]
- Gatt K, Schembri J, Katsanos KH, et al. Inflammatory Bowel Disease [IBD] and Physical Activity: A Study on the Impact of Diagnosis on the Level of Exercise Amongst Patients With IBD. J Crohns Colitis 2019;13:686-92. [Crossref] [PubMed]
- Eckert KG, Abbasi-Neureither I, Köppel M, et al. Structured physical activity interventions as a complementary therapy for patients with inflammatory bowel disease - a scoping review and practical implications. BMC Gastroenterol 2019;19:115. [Crossref] [PubMed]
- van Erp LW, Roosenboom B, Komdeur P, et al. Improvement of Fatigue and Quality of Life in Patients with Quiescent Inflammatory Bowel Disease Following a Personalized Exercise Program. Dig Dis Sci 2021;66:597-604. [Crossref] [PubMed]
- Gracie DJ, Hamlin PJ, Ford AC. The influence of the brain-gut axis in inflammatory bowel disease and possible implications for treatment. Lancet Gastroenterol Hepatol 2019;4:632-42. [Crossref] [PubMed]
- Sun Y, Li L, Xie R, et al. Stress Triggers Flare of Inflammatory Bowel Disease in Children and Adults. Front Pediatr 2019;7:432. [Crossref] [PubMed]
- Bisgaard TH, Allin KH, Keefer L, et al. Depression and anxiety in inflammatory bowel disease: epidemiology, mechanisms and treatment. Nat Rev Gastroenterol Hepatol 2022;19:717-26. [Crossref] [PubMed]
- Kaplan GG, Hubbard J, Korzenik J, et al. The inflammatory bowel diseases and ambient air pollution: a novel association. Am J Gastroenterol 2010;105:2412-9. [Crossref] [PubMed]
- Opstelten JL, Beelen RMJ, Leenders M, et al. Exposure to Ambient Air Pollution and the Risk of Inflammatory Bowel Disease: A European Nested Case-Control Study. Dig Dis Sci 2016;61:2963-71. [Crossref] [PubMed]
- Shaw SY, Blanchard JF, Bernstein CN. Association between the use of antibiotics in the first year of life and pediatric inflammatory bowel disease. Am J Gastroenterol 2010;105:2687-92. [Crossref] [PubMed]
- Theochari NA, Stefanopoulos A, Mylonas KS, et al. Antibiotics exposure and risk of inflammatory bowel disease: a systematic review. Scand J Gastroenterol 2018;53:1-7. [Crossref] [PubMed]
- Nguyen LH, Örtqvist AK, Cao Y, et al. Antibiotic use and the development of inflammatory bowel disease: a national case-control study in Sweden. Lancet Gastroenterol Hepatol 2020;5:986-95. [Crossref] [PubMed]
- Navarro E, Funtikova AN, Fíto M, et al. Prenatal nutrition and the risk of adult obesity: Long-term effects of nutrition on epigenetic mechanisms regulating gene expression. J Nutr Biochem 2017;39:1-14. [Crossref] [PubMed]
- Ventham NT, Kennedy NA, Nimmo ER, et al. Beyond gene discovery in inflammatory bowel disease: the emerging role of epigenetics. Gastroenterology 2013;145:293-308. [Crossref] [PubMed]
- Relton CL, Davey Smith G. Epigenetic epidemiology of common complex disease: prospects for prediction, prevention, and treatment. PLoS Med 2010;7:e1000356. [Crossref] [PubMed]
- Cheung P, Vallania F, Warsinske HC, et al. Single-Cell Chromatin Modification Profiling Reveals Increased Epigenetic Variations with Aging. Cell 2018;173:1385-1397.e14. [Crossref] [PubMed]
- Ventham NT, Kennedy NA, Adams AT, et al. Integrative epigenome-wide analysis demonstrates that DNA methylation may mediate genetic risk in inflammatory bowel disease. Nat Commun 2016;7:13507. [Crossref] [PubMed]
- Waddington CH. The epigenotype. 1942. Int J Epidemiol 2012;41:10-3. [Crossref] [PubMed]
- Jenke AC, Zilbauer M. Epigenetics in inflammatory bowel disease. Curr Opin Gastroenterol 2012;28:577-84. [Crossref] [PubMed]
- Allis CD, Jenuwein T. The molecular hallmarks of epigenetic control. Nat Rev Genet 2016;17:487-500. [Crossref] [PubMed]
- Zhu J, Adli M, Zou JY, et al. Genome-wide chromatin state transitions associated with developmental and environmental cues. Cell 2013;152:642-54. [Crossref] [PubMed]
- Fellows R, Varga-Weisz P. Chromatin dynamics and histone modifications in intestinal microbiota-host crosstalk. Mol Metab 2020;38:100925. [Crossref] [PubMed]
- Wigler M, Levy D, Perucho M. The somatic replication of DNA methylation. Cell 1981;24:33-40. [Crossref] [PubMed]
- Margueron R, Reinberg D. Chromatin structure and the inheritance of epigenetic information. Nat Rev Genet 2010;11:285-96. [Crossref] [PubMed]
- G N. Zilbauer M. Epigenetics in IBD: a conceptual framework for disease pathogenesis. Frontline Gastroenterol 2022;13:e22-7. [Crossref] [PubMed]
- McDermott E, Ryan EJ, Tosetto M, et al. DNA Methylation Profiling in Inflammatory Bowel Disease Provides New Insights into Disease Pathogenesis. J Crohns Colitis 2016;10:77-86. [Crossref] [PubMed]
- Somineni HK, Venkateswaran S, Kilaru V, et al. Blood-Derived DNA Methylation Signatures of Crohn's Disease and Severity of Intestinal Inflammation. Gastroenterology 2019;156:2254-2265.e3. [Crossref] [PubMed]
- Rajamäki K, Taira A, Katainen R, et al. Genetic and Epigenetic Characteristics of Inflammatory Bowel Disease-Associated Colorectal Cancer. Gastroenterology 2021;161:592-607. [Crossref] [PubMed]
- Lloyd-Price J, Arze C, Ananthakrishnan AN, et al. Multi-omics of the gut microbial ecosystem in inflammatory bowel diseases. Nature 2019;569:655-62. [Crossref] [PubMed]
- Kelly D, Kotliar M, Woo V, et al. Microbiota-sensitive epigenetic signature predicts inflammation in Crohn's disease. JCI Insight 2018;3:e122104. [Crossref] [PubMed]
- Catalanotto C, Cogoni C, Zardo G. MicroRNA in Control of Gene Expression: An Overview of Nuclear Functions. Int J Mol Sci 2016;17:1712. [Crossref] [PubMed]
- Fisher K, Lin J. MicroRNA in inflammatory bowel disease. New Insights into Inflammatory Bowel Disease. IntechOpen, 2016.
- Friedman RC, Farh KK, Burge CB, et al. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res 2009;19:92-105. [Crossref] [PubMed]
- McKenna LB, Schug J, Vourekas A, et al. MicroRNAs control intestinal epithelial differentiation, architecture, and barrier function. Gastroenterology 2010;139:1654-64, 1664.e1.
- Adams AT, Kennedy NA, Hansen R, et al. Two-stage genome-wide methylation profiling in childhood-onset Crohn's Disease implicates epigenetic alterations at the VMP1/MIR21 and HLA loci. Inflamm Bowel Dis 2014;20:1784-93. [Crossref] [PubMed]
- Shi C, Liang Y, Yang J, et al. MicroRNA-21 knockout improve the survival rate in DSS induced fatal colitis through protecting against inflammation and tissue injury. PLoS One 2013;8:e66814. [Crossref] [PubMed]
- Chen Y, Xiao Y, Ge W, et al. miR-200b inhibits TGF-β1-induced epithelial-mesenchymal transition and promotes growth of intestinal epithelial cells. Cell Death Dis 2013;4:e541. [Crossref] [PubMed]
- Del Follo-Martinez A, Banerjee N, Li X, et al. Resveratrol and quercetin in combination have anticancer activity in colon cancer cells and repress oncogenic microRNA-27a. Nutr Cancer 2013;65:494-504. [Crossref] [PubMed]
- Biersack B. Current state of phenolic and terpenoidal dietary factors and natural products as non-coding RNA/microRNA modulators for improved cancer therapy and prevention. Noncoding RNA Res 2016;1:12-34. [Crossref] [PubMed]
- Ho SM, Lewis JD, Mayer EA, et al. Challenges in IBD Research: Environmental Triggers. Inflamm Bowel Dis 2019;25:S13-23. [Crossref] [PubMed]
Cite this article as: Rodrigues BAG, Steigleder KM, Menta PLR, de Castro MM, Milanski M, Leal RF. The exposome-diet-epigenome axis in inflammatory bowel diseases—a narrative review. Dig Med Res 2024;7:6.


