
Narrative review of gut microbiota and liver diseases: facts and fictions
Introduction
The gastrointestinal tract is colonized by a wide range of commensal microorganisms, approximately 100 billion (1) that play an important role in maintaining health. This intestinal microbiota represents 10 times more than the total number of cells in the human body. It is made up of bacteria, viruses and fungi, which are divided into 6 groups: Gram negative Bacteroidetes, Proteobacteria, Gram positive Firmicutes and Acinetobacteria, Fusobacteria and Verrucomicrobia (2). For all the genetic material of the microbiota, it has been called the second genome contained in the human body (1). The intestinal microbiota plays a key role in maintaining health condition and drives diverse immune, nutritional and metabolic functions within the host. It’s composition and diversity very according to different factors such as the form of birth (caesarean section or vaginal route), maternal microbiota, infant diet (breast milk or formula), geographic location, age, gender (3), diet in adulthood (meat-based or vegetarian/vegan), in addition to the use of antibiotics (2). The close relationship between the gut microbiota and the liver, formerly known as gut-liver axis, is widely studied, accepted but not fully understood (4). The alteration of the gut-liver axis due to intestinal dysbiosis, intestinal delayed transit, bacterial overgrowth of the small intestine (5) and increased intestinal permeability seen in chronic liver disease (CLD) provoke a chronic systemic inflammatory state, which drives the progression of CLDs (6). As shown in Table 1 (7-9).
Table 1
| Clinical condition | Phylum | Species |
|---|---|---|
| Normal | Firmicutes, Bacteroidetes, Verrucombia Actinobacteria, Fusobacteria, Proteobacteria | Lactobacillus, Clostridium, Enterococcus, Ruminococcus, Bacteroides, Prevotella |
| Obesity | Firmicutes (↑), Mitsuokella (↑), Bacteroidetes (↓), Bifidobacterium (↓), Odoribacter (↓), Phascolartobacterium (↓), Oscillospira (↓) | Akkermansia muciniphila (↓), Tuminococcaceae, Rikenellaceae, Mollicutes (↑), Alistipes (↑), Bilophila (↑), Roseburia, Eubacterium rectale (↓), Rominococcus bromii (↓) |
| Diabetes | T2DM: Firmicutes (↑), Lactobacillus (↓) Bacteroidetes (↓) | T2DM: Roseburia (↓), Faecalibacterium prauznitzii (↓), E. coli (↑), Ruminococcus (↓), Akkernansia muciniphila (↓), Clostridium (↓) |
| T1DM: Parabacteroides | T1DM: Parabacteroides (↓), Akkermansia muciniphila (↓), E. coli (↑) | |
| MAFLD | Acidaminococcus (↑), Akkermansia (↑), Allisonella (↑), Anaerococcus (↑), Bradyrhizobium (↑), Dorea (↑), Escherichia (↑), Parabacteroides (↑), Robinsonella (↑), Alistipes (↓) Anaerosporobacter (↓), Coprobacter (↓), Moryella (↓), Subdoligranulum (↓) | Veillonella (↑), E. lenta (↑), C. clostridioforme (↑), C. ramosum (↑), Lachnospiraceae bacterium (↑), Barnesiella intestinihominis (↑), Faecalibacterium prausnitzii (↓), E. rectale (↓), C. coccoides (↓) |
| Cancer | Firmicutes (↓), Proteobacteria | Bilophila wadsworthia, Fusobacterium nucleatum (↑) |
MAFLD, metabolic associated fatty liver disease.
We present the following article in accordance with the Narrative Review reporting checklist (available at https://dmr.amegroups.com/article/view/10.21037/dmr-21-85/rc).
Methods
A comprehensive research of Medline In-Process and other Non-Indexed Citations was conducted, using MEDLINE by the US National Library of Medicine National Institutes of Health and Google Scholar (prior to October 31, 2021) to retrieve relevant articles. Literature search was made using medical subject headings (MeSH) and text words related to microbiota and chronic liver disease. Controlled terms were used to search for studies (Chronic Liver disease [ALL] AND Microbiota), (Microbiota [ALL] AND bile acids), (microbiota [ALL] AND intestinal barrier), (Microbiota [ALL] AND metabolic disturbance), (Microbiota [ALL] AND systemic inflammation), (microbiota [ALL] autoimmune hepatitis, primary biliary cirrhosis, primary sclerosing cholangitis), (microbiota [ALL] AND hepatic encephalopathy), (microbiota [ALL] AND Non-Alcoholic Fatty Liver Disease AND Alcoholic Liver Disease), (Microbiota [ALL] AND Hepatocellular carcinoma), (Microbiota [ALL] AND Acute-on-Chronic liver failure). Additional studies were located by manual search using references from retrieved articles. Research strategies were shown in Table 2.
Table 2
| Items | Specification |
|---|---|
| Date of search (specified to date, month and year) | From October 1st to 31st 2021 |
| Databases and other sources searched | Pubmed/Medline, Google Scholar and Cochrane |
| Search terms used (including MeSH and free text search terms and filters) | Controlled terms were used to search for studies (Chronic Liver disease [ALL] AND Microbiota), (Microbiota [ALL] AND bile acids), (microbiota [ALL] AND intestinal barrier), (Microbiota [ALL] AND metabolic disturbance), (Microbiota [ALL] AND systemic inflammation), (microbiota [ALL] autoimmune hepatitis, primary biliary cirrhosis, primary sclerosing cholangitis), (microbiota [ALL] AND hepatic encephalopathy), (microbiota [ALL] AND Non-Alcoholic Fatty Liver Disease AND Alcoholic Liver Disease), (Microbiota [ALL] AND Hepatocellular carcinoma), (Microbiota [ALL] AND Acute-on-Chronic liver failure). Additional studies were located by manual search using references from retrieved articles |
| Timeframe | 1995–2021 |
| Inclusion and exclusion criteria (study type, language restrictions etc.) | Research studies, randomized or non randomized trials and reviews/meta-analysis. We excluded studies that were not in English or Spanish Language, or that were published before 1995. We excluded citations that were not completely related to the topic or that did not have an abstract available or that we do not have access to the full text. A total of 200 articles were obtained. We decided to include 80 articles that fulfilled the inclusion criteria |
| Selection process (who conducted the selection, whether it was conducted independently, how consensus was obtained, etc.) | The research was performed by the main author, the selection was confirmed by all the authors of the manuscript |
| Any additional considerations, if applicable | None |
Intestinal mucosal immune barrier: first line of defense
The intestinal mucosa is constantly changing, with exchange of nutrients, ions, and fluids through the epithelium, but it is also in contact with many potentially pathogenic antigens and microorganisms. It’s architecture is so elaborate that it has a contact surface of approximately 300–400 m2. It is colonized by bacteria in a range of 1×104 cells in the small bowel to 1×1014 in the colon (10). The intestinal immune system is made up of 3 different structures: Peyer’s patches, lamina propria, and the epithelium (mucosa-associated lymphoid tissue, MALT) (11). The mucosal layer produces glycoprotein, the main source of nutrition for some bacterial species, it also represents the main source of immunoglobulin A (IgA) and antimicrobial peptides, this helps to preserve intestinal integrity that protects against bacterial translocation. Another important anatomical component of the intestinal barrier is the tight junction (TJ) proteins that holds epithelial cells together while preserving the integrity of the intestinal barrier; when these proteins are under-expressed, an increase in the passage of lipopolysaccharide (LPS) and bacterial products from the bowel to the portal vein is observed (12). The second line of defense is the immune cells present in the intestinal barrier, composed mainly of lymphocytes, natural killer T cells, mononuclear phagocytes and dendritic cells. The presence of these immune cells is important to preserve intestinal homeostasis, avoiding an increase in intestinal permeability and the activation of inflammatory pathways (13) (Figure 1).
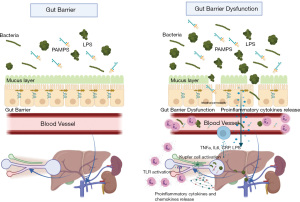
Anatomic barriers of the mucosa
The anatomic barriers of the mucosa are composed by:
- Extracellular components: most of the mucosal surface is hydrated by a gel layer formed by mucin, which is secreted by specialized epithelial cells such as gastric and intestinal foveolar mucous cells and goblet cells. This creates a barrier that prevents most bacteria from coming into direct contact (14). The importance of this is shown in pathologies such as cystic fibrosis, in which the hyperviscous production of mucus contributes to infectious pulmonary, pancreatic and intestinal pathologies (15).
- Cellular components: made up of specialized epithelial cells, which are interconnected by junctions and are covered by mucus and bactericidal peptides. This allows a small number of bacteria to penetrate the intestinal epithelium; however, there are many physical and pharmacological agents that can harm it (16). The microbiota and the intestinal mucosa are in constant interaction to maintain homeostasis. When the balance is broken, it predisposes to different diseases (11).
Interplay between the gut microbiome and the liver
The relationship between the microbiota and the progression of CLD is continuously investigated, constantly evolving and not completely understood, extensive scientific evidence is still missing to fully understand the importance of the microbiota and liver disease. Current data have shown that patients with liver cirrhosis have increased bacteremia and increased levels of lipopolysaccharides, as well as increased intestinal permeability (6). Primary human bile acids, cholic acid (CA) and chenodeoxycholic acid (CDCA), are synthesized in the liver, secreted into the intestine, and undergo bacterial biotransformation to generate secondary bile acids, lithocholic acid (LCA) and deoxycholic acid (DCA) by the bile salt hydrolase (BSH) (17). The circulation of secondary bile acids from the intestine to the liver activates the farnesoid X receptor (FXR) that suppresses the production of bile acid. Intestinal FXR seems to modulate gut microbiota by inducing Acetatifactor (Clostridium cluster XIV family) and Bacteroides to convert primary bile acids into secondary bile acids that activate takeda G coupled receptor 5 (TGR5) which in turn stimulates the secretion of glucagon like peptide 1 (GLP1) improving hepatic metabolism and insulin sensitivity (18,19). The primary classes of such receptors include the Toll-like (TLR) and NOD-like (NLR) receptors that recognize a variety of bacterial products, including LPS, flagellin, peptidoglycan, and bacterial DNA. The main consequence of TLR/NLR detecting its cognate agonists is to widely induce host-defense gene expression that can protect against numerous microbes. Numerous studies indicate that, like most macrophages populations, Kupffer cells respond to very low concentration of LPS by activating NF-kB and producing pro-inflammatory cytokines, suggesting that these cells would respond at physiologically relevant levels of microbial products that reach the liver (20).
New insights on gut microbiota in liver disease
The liver has a double blood supply, almost 70% of its blood comes from the intestines through the portal circulation and the rest through the hepatic artery. Although the intestinal mucosa acts as an effective barrier against the translocation of microbes and microbial products from the gut to the circulation, small amounts of these enter the portal venous blood. The liver, strategically located between highly contaminated bowel and sterile systemic circulation, functions as a filter. The proposed mechanisms of gut–liver interaction in these diseases include alterations in composition of the gut microbiota, bacterial overgrowth of the small intestine, increased permeability of the small bowel and alterations in mucosal and systemic immunity. With the development of better technology, such as genetic sequencing analysis, it is possible to classify bacteria based on DNA sequence. It has been recognized that short-chain fatty acids such as butyric acid, acetic (microbiota fermentation) can decrease the T-cell-mediated immune response through an epigenetic mechanism (17,21). Probiotics have also been used in murine hepatocellular carcinoma (HCC) models to oppose dysbiosis and have shown a reduction in HCC development (17).
Although research has traditionally focused on the bacterial component of the gut microbiota, research of enteric fungal and viral species has increased due to advances in sequencing and analysis technologies (21,22).
Microbiota and metabolic disturbance
Dysbiosis has been associated with important metabolic consequences, such as increased expression of genes that encode enzymes related to carbohydrate metabolism, closely related to the development of metabolic associated fatty liver disease (MAFLD) (23).
Diabetes mellitus type 2
Alterations in glucose metabolism (glucose intolerance and type 2 diabetes mellitus) represent pathologies with a high prevalence; in 2012 it was estimated that approximately 9.3% (29.1 million people) in the United States have type 2 diabetes mellitus. The role of dysbiosis in the intestinal microbiota has recently been investigated, which could play an important role in pathogenesis. Different observational studies have described differences between the microbiota of people with type 2 diabetes mellitus versus healthy people (24). Opportunistic pathogens are frequently described in T2DM microbiota communities, including the species Clostridium ramosum, hathewayi and symbiosum, Bacteroides caccae, Eggerthella lenta and Butyrate-producing microbes are particularly depleted specifically in the Clostridiales order, including the genera Ruminococcus and Subdoligranulum, and the species Eubacterium rectale, Faecali prausnitzii, Roseburia intestinalis and Roseburia inulinivorans, pre-diabetic patients also demonstrate similar findings in their microbiota communities, including a decrease in microbial diversity; depletion in the numbers of the genera Akkermansia and Clostridium; and increase in Ruminococcus and Streptococcus (25).
There are different mechanisms that have been proposed through which the microbiota alters glucose metabolism:
- Production of metabolites: short chain fatty acids (SCFAs; butyrate mainly produced mainly by Firmicutes and acetate produced by Bacteroidetes) are the main products resulting from anaerobic fermentation of dietary fiber and have a direct impact on the hepatic metabolism of glucose, decreasing glycolysis and gluconeogenesis, increasing glycogen synthesis.
- Stimulation of the GPR41 receptor by butyrate and propionate can induce intestinal gluconeogenesis, promoting the secretion of the incretin GLP-1 (25). Better HbA1c control (<7%) has been described in patients who receive fiber in the diet compared to the control group, as well as the consumption of a Mediterranean diet that improves glucose sensitivity in patients with high cardiovascular risk. This is due to the production of large amounts of succinate by bacterial fermentation of dietary fiber which improves glycemic control through the activation of intestinal gluconeogenesis (26,27).
Type 1 diabetes mellitus
It is an autoimmune disease in which pancreatic destruction of B cells causes insulin deficiency resulting in hyperglycemia and risk of ketoacidosis. The presence of 5 bacterial genera has been identified with the development of type 1 diabetes mellitus, mainly Parabacteroides. A decrease in the production pathways of short-chain fatty acids such as butyrate has been documented in patients who develop antibodies against pancreatic islets (28,29).
Microbiota and cardiovascular risk
Many researchers have reported the association between cardiovascular risk and the microbiota. Bacterial DNA has been detected in atherosclerotic plaques. Elevated levels of lipopolysaccharides produced by the bacterial wall of the microbiota have been linked to modulation of inflammation, immunity and vascular function. Patients with decompensated heart failure have higher blood endotoxin levels compared to stable patients (30,31). The strongest evidence for the relationship between the microbiota and cardiovascular risk is related to the microbial metabolism of dietary factors such as carnitine and choline. These metabolites are potentially the cause of atherosclerosis. Trimethylamine is produced by the microbiota from nutrients containing L-carnitine, which promotes the formation of foam cells and atheroma plaques in animal models (32). Many microorganisms such as Chlamydophila pneumoniae, Porphyromonas gingivalis, Helicobacter pylori, Influenza A virus, Hepatitis C virus, cytomegalovirus, and human immunodeficiency virus have been associated with an increased risk of cardiovascular disease. Infections contribute to atherosclerosis through two predominant mechanisms: direct infection of the blood vessel wall (making it prone to plaque formation), or indirectly with an infection at a distant site by promoting proinflammatory mediators of a systemic immune response that predispose to plaque growth (33).
Microbiota and systemic inflammation
The microbiota has a variety of functions that play a role in its ability to grow and colonize. Related mechanisms have been proposed between the microbiota and inflammatory markers, among them:
- Lipopolysaccharides, also known as endotoxins, are found as a component of the cell wall of Gram-negative bacteria. Its increased values have been reported in obesity, metabolic disorders, tissue inflammation and pancreatic dysfunction (34). Under normal conditions, the intestinal barrier can minimize the translocation of LPS from the intestinal lumen to the circulation. Due to the dysfunction of the intestinal barrier by diet, the microbiota allows the movement of LPS producing a systemic immune response mediated by Toll receptors (TLR 4) (35).
- SCFAs. Bacteria can metabolize complex carbohydrates to SCFAs (30,31), SCFAs play an important role in reducing the activation of the inflammatory cascade. Among them, butyrate is an anti-inflammatory metabolite, which reduces the risk of developing insulin resistance, it also reduces the translocation of LPS. Faecalibacterium prausnitzii was identified as a butyrate-producing bacterium, a significant decrease of this microorganism has been documented in obese individuals (34).
- C-reactive protein (CRP). It is an acute phase reactant, associated with cardiovascular disease, obesity and type 2 diabetes. The genus Phascolarctobacterium has been associated with low levels of CRP, this relationship may explain why the decline of the genus has been associated with inflammation of the colon. Phascolarctobacterium are producers of propionate, a short-chain fatty acid that inhibits the inflammatory response. Faecalibacterium levels are inversely correlated with CRP levels (34).
Effect of microbiota in CLD progression and complications
Alcoholic liver disease (ALD)
Alcohol-related CLDs are responsible for 0.9% of all deaths and 47.9% of all deaths attributable to cirrhosis. Chronic alcohol intake allows intestinal bacteria overgrowth and dysbiosis in humans and animals. In humans, it is defined as the growth of at least 1×105 in a bean aspirate culture. Chronic alcohol consumption decreases intestinal levels of antimicrobial-regenerating islet-derived (REG)-3 lectins. Specific intestinal deficiency in Reg3b or Reg3g increases the number of intestinal bacteria and their translocation to mesenteric lymph nodes and the liver causing the development and progression of ethanol-induced steatohepatitis. In contrast, Reg3g overexpression in the intestinal epithelium protects animals from ethanol-induced liver injury (36). Patients with ALD have dysbiosis, represented by a reduction of the mean abundances of Bacteroidetes and an increase in Proteobacteria. Patients with alcohol-related liver cirrhosis have been documented to have a reduction in species as Lachnospiraceae, Lactobacillus spp, Bifidobacterium spp, Ruminococcaceae, Clostridiales XIV, and an increase in Enterobacteriaceae, including the predominant genus Escherichia coli, Bacteroidaceae (37). Gut bacteria have also been reported to differ between stages of ALD and are associated with its progression. Streptococcus have been associated with the severity and progression of ALD (38). Bacteria of the Lactobacillus and Veillonella genera have also been reported to increase in patients with alcoholic hepatitis (AH), mainly in severe cases; on the other hand, bacteria as Eubacterium, Oscillibacter, Prevotella, Prevotellaceae and Clostridiales are reduced in patients with severe AH (39).
MAFLD
It is considered one of the main causes of CLD in the general population. It is a disease with a wide clinical spectrum that ranges from steatohepatitis, fibrosis, cirrhosis and/or HCC. The most serious form is steatohepatitis and it can progress to cirrhosis in 20% of patients (40). The profile of microorganisms present in fatty liver disease is frequently associated with obesity and excess abdominal adiposity, there is a decrease in diversity compared to healthy people (41). An increase in the radius of Firmicutes/Bacteroidetes is reported, with an increase in Bacteroidetes and a decrease in Firmicutes. Numerous pathophysiological mechanisms have attempted to explain the relationship between the microbiota and liver-systemic inflammation. The importance of increase in intestinal permeability has been described, causing with the continuous passage of LPS and pathogen associated molecular patterns (PAMPS) through the portal vein causing endotoxemia and immune dysregulation, together with the bacterial overgrowth (SIBO) that occurs (42). All of the aforementioned causes the activation of inflammatory pathways with the activation of Toll-like receptors and the secretion of cytokines, interleukin 8 (IL8), IL6 and TNFα, enhancing liver inflammation (steatohepatitis), fibrosis and systemic inflammation (43) (Figure 2).
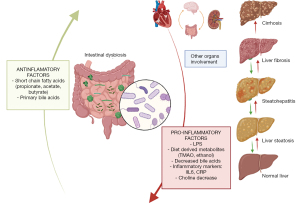
Endogenous ethanol (EnEth) producing bacteria have been shown to play a role in the onset and exacerbation of various stages of MAFLD (8). EnEth production is increased, and the prevalence of intestinal permeability and bacterial translocation is higher, the Klebsiella pneumoniae strain (W14) with high alcohol production (HiAlc) was isolated from the feces of a patient with severe non-alcoholic steatohepatitis. W14 was considered the potential culprit for MAFLD, which leads to hepatic steatosis and mitochondrial dysfunction (44). Studies have shown that Enterobacteriaceae, Proteobacteria, and Escherichia were the only phylum, family, and genus types exhibiting significant differences in obese subjects with or without steatohepatitis, and steatohepatitis microbiomes exhibited a lower percentage of Bacteroidetes (36). Interestingly, MAFLD patients have increased concentrations of Bacteroides and Ruminococcus and a reduction of Prevotella mainly in advanced disease.
Autoimmune diseases
Autoimmune hepatitis (AIH)
Some studies have characterized the microbiome of patients who had not received steroid treatment for AIH. Wei et al. found that these patients had a depletion of obligate anaerobes such as Faecalibacterium, as well as the expansion of Veillonella. Veillonella disbar was linked to the severity of the disease and inflammation of the liver (45). A disease-specific decrease in the relative abundance of Bifidobacterium in patients with AIH have been described. In other study by Liwinski et al., found that the lack of Bifidobacterium was associated with failure to achieve AIH remission and with an increased disease activity (46).
Primary biliary cholangitis (PBC)
PBC is the most common autoimmune liver disease, occurring approximately 0.1% of women in their 40s. It is characterized by the presence of antimitochondrial antibodies and the destruction of the intrahepatic biliary ducts, allowing fibrosis and potentially cirrhosis. The microbiota plays an important role, a decrease in genera such as Faecalibacterium, Bacteroides, Sutterella and Oscillospira spp has been described, as well as an increase in Enterobacteriaceae such as E. coli, K. pneumonia and E. cloacae (47). Lipoteichoic acid, a cell wall component of gram-positive bacteria, was augmented in destroyed bile ducts of liver samples and in serum of PBC patients compared to patients with hepatitis C (48).
Primary sclerosing cholangitis (PSC)
PSC is a cholestatic liver disease, characterized by inflammation and stenosis of the bile ducts. Recent studies have described the relationship of this disease with the gut- liver axis, with a decreased microbiota diversity in patients with PSC. Characterization of the fecal microbiota in adult PSC has shown elevated levels of certain species, such as Veillonella, Fusobacterium, Streptococcus parasanguinis, Enterococcus faecium, E. coli, Faecalibacterium, Ruminococcus and Roseburia. There is also a reduction in short-chain fatty acid-producing bacteria, such as Firmicutes, Faecalibacterium, and Coprococcus, as well as the reduction of Anaerostipes hadrus, Proteobacteroides distasonis and Blautia obeum (49-51). Veillonella has also been associated with other chronic inflammatory disorders. It appears that the abundance of Veillonella species and the over-representation of the genus Enterococcus exhibit a signature in PSC patients. This suggests that Veillonella and Streptococcus species might be related to the immune modulation and the activation of inflammatory pathways in PSC (51). Interestingly, the microbiota in PSC patients seems to be similar, whether or not they have inflammatory bowel disease (IBD); suggesting that PSC might drive microbiota alterations (52).
End stage CLD and intestinal microbiome
Cirrhosis, the final stage of liver fibrosis, is classified into compensated o decompensated states. The main complications such as hepatic encephalopathy and spontaneous bacterial peritonitis are related to alterations in the gut- liver axis (19). There are changes in the composition of the microbiota between healthy control and those with liver cirrhosis. The main changes observed are the reduction in Lachnospiraceae and Ruminococcaceae, bacteria that are associated with the production of SCFAs and the formation of primary and secondary bile acids. An increase in pathogens such as Enterobacteriaceae that produce endotoxins and lipopolysaccharides, has also been observed, reducing SCFAs, causing an increased intestinal permeability and promoting the passage of LPS and bacterial products to the liver; causing hepatic and systemic inflammation (53). The increase of bacteria of oral origin, such as Veillonella parvula and Streptococcus salivarius, has been shown in cirrhotic patients who received proton pump inhibitors (PPIs), increasing intestinal permeability and promoting cirrhosis progression (54). Other studies have shown an increase in the microbiota of the oral cavity such as Veillonellaceae, Porphyromonadaceae and Streptococcaceae, later it was reported that this is an epiphenomenon due to the use of PPIs, which decreases when they are suspended (55). Other oral-derived bacteria Rothia, Streptococcaceae and Veillonellaceae have been associated with cirrhosis complications and progression (56,57). Changes in the concentration of colonic fungi (Candida spp) have been described in patients with advanced CLD. This is compounded using broad-spectrum antibiotics. These findings indicate that cirrhosis is a systemic disease that worsens with dysbiosis. Importantly, the concentration of potentially pathogenic bacteria, such as Enterobacteriaceae and Streptococcaceae, associated with the reduction of beneficial populations such as Lachnospiraceae could accelerate the progression and affect the prognosis of these patients. Intestinal dysbiosis seem to be more severe the more advanced the hepatic decompensation is (36).
Cirrhosis complications and microbiome
Hepatic encephalopathy and microbiota
Hepatic encephalopathy is a complication of cirrhosis, it is a syndrome characterized by neurocognitive deterioration, where the pathophysiological substrate is hyperammonemia produced mainly by Enterobacteria (19). Hepatic encephalopathy is a neurological complication that occurs in individuals with CLDs typically divided into three types [type A resulting from acute hepatic failure (ALF), type B resulting from portosystemic bypass or shunting, and type C resulting from cirrhosis]. Ammonia has been known to play a central role in the pathogenesis of HE. Urease is a bacterial enzyme that catalyzes the hydrolysis of urea to carbamate and ammonia. Compared to healthy controls, intestinal microbiota on patients with cirrhosis have an abundance of 75,245 genes according to quantitative metagenomics. The genus Bacteroidetes decreases according with a decrease in liver function. Some intestinal microbiota have been correlated with the pathological mechanisms, processes and outcomes of HE. The translocation of Methylobacterium extorquens and Stenotrophomonas pavanii into the peripheral blood system enhances the risk of HE.
Effects of clinical treatment on the intestinal metabolome in hepatic encephalopathy
Various drugs are used to lower blood ammonia levels. Lactulose is the most widely used ammonia-reducing drug. It is a laxative drug, nonabsorbable disaccharide. It also inhibits intestinal bacterial overgrowth, translocation and intestinal resistance, in patients with liver cirrhosis, clinical doses of lactulose promote the growth of beneficial bacteria, such as Lactobacillus and Bifidobacterium, increase the abundance of hydrogen‐producing bacteria Rikenellaceae and Prevotellaceae, and mucin‐degrading bacteria Akkermansia and Helicobacter; and decrease the abundance of harmful bacteria Desulfovibrionaceae (58,59). Intestinal antibiotics have been tested for the treatment of HE. Among them the most widely used is Rifaximin is a non-systemic antibiotic approved for the treatment of HE. It has bactericidal and bacteriostatic activity in vitro against aerobic and anaerobic Gram-positive and negative species, and can also reduce bacterial virulence and translocation, it also inhibits bacterial adherence to the intestinal mucosal, with a decrease in Roseburia, Haemophilus, Veillonella and Streptococcus and increase in Lactobacilli after 10 days of rifaximin administration, this is in line with the evidence that rifaximin is able to promote the growth of beneficial bacteria (60). Other antibiotics such as neomycin are not recommended because of their side effects (58). Probiotics (including Bifidobacterium longum, Lactobacillus paracasei, Bifidobacterium infantis, Lactobacillus acidophilus, Bifidobacterium breve, Lactobacillus plantarum, Lactobacillus bulgaricus, and Streptococcus thermophilus) have several mechanisms that may be involved in hepatic encephalopathy improvement and may include: modulation of the gut microbiota, reduce total ammonia in portal blood by decreasing bacterial urease activity, decrease ammonia absorption by lowering pH, decrease intestinal permeability, and improve nutritional status of gut epithelium, in addition, probiotics can stimulate the production of SFCAs by increasing the abundance of multiple bacteria, including Syntrophococcus sucromutans, Calibacterium prausnitzii, and Alistipes shahii (61,62).
HCC and microbiota
The persistent inflammation and liver fibrosis associated with CLD described above also tend to favor the accumulation of genetic and epigenetic changes, such as mutations in the telomerase reverse transcriptase promoter, tumor protein p53 or signaling genes, which lead to HCC development. More recent studies have delineated that the gut microbiota in NAFLD-related HCC animal models has increased Bifidobacterium and a group of organisms in the Clostridiales order, formerly known as the “Clostridium cluster XVIII.” Both groups are involved in bile acids (BA) modification and are involved in the conversion of primary BAs to secondary BAs, such as deoxycholic acid (DCA) (17). No differences have been found in the intestinal microbiota of patients who developed or had previous HCC; however, specific microbiota traits have been described in patients with HCC. In HCC NAFLD patients, a higher concentration of Enterococcus, Bacteroides, Clostridium, Paraprevotella and Ruminococcaceae, and a reduction of Bifidobacterium, Alphaproteobacteria and Akkermansia have been described (57,60). Streptococcus and Enterococcus have been reported to be related to the progression of cirrhosis and the development of HCC. (60) Lapidot et al. demonstrated that NAFLD patients who developed HCC had increased concentrations of Akkermansia (63). It has also been reported that HCC patients have a reduction in bacteria that produce SFCAs as Roseburia, Odoribacter, Clostridium sensu stricto (64). The Clostridial Anaerotruncus and Clostridium sensu strictus taxa have also been reported to be associated with the absence of HCC. Yoshimoto et al. found an association between HCC and the predominantly Clostridium XI and XIVa clusters (65).
Acute on chronic liver failure and microbiome (ACLF)
Acute on chronic liver failure is a clinical entity that occurs in patients with CLD with a triggering event that decompensates it. It is also characterized by multiple organ failure, including hepatic encephalopathy, has a rapid progression, a poor prognosis and a high mortality. The microbiota presents classic alterations such as the increase in Streptococcaceae, Enterobacteriaceae, Alcaligenaceae and Porphyromonadaceae, Lachnospiraceae, and a reduction in Bacteroidetes, Proteobacteria, Firmicutes (66). There is evidence that translocation of components of the gut microbiota (LPS), facilitated by different pathogenic mechanisms (increased intestinal permeability and portal hypertension), is an important driver of decompensation by the induction of systemic inflammation, and thereby ACLF, as shown in Table 3 (49,67-69).
Table 3
| Liver disease | Intestinal microbiome |
|---|---|
| MAFLD | Increase: Bacteroidetes, Clostridium coccoides, Bacteroides, E. coli, Bacteroides vulgatus, Acidaminococcus, Streptococcus, Gallibacterium, enterobacteriaceae, Ruminococcaceae (Ruminococcus gnavus), Enterococcus, Phascolarctobacterium, Blautia, Veillonella parvula, Veillonella atypica. Decrease: Firmicutes, Clostridiales, Faecalibacterium, Prevotella, Eubacterium eligens, E. rectale, Facalibacterium prausnitzii, Bifidobacterium, Blautia |
| ALD | Increase: Enterobacteriaceae, including the predominant species Escherichia coli, Bacteroidaceae. Decrease: Lachnospiraceae, Lactobacillus spp, Bifidobacterium spp, Ruminococcaceae, Clostridiales XIV |
| Autoimmune (PSC, PBC, AIH) | PSC Increase: Bacilli, Pseudomonas, Streptococcus, E. coli, Barnesiellaceae, Blautia, Ruminococcus obeum, Veillonella, Lactobacillus. Decrease: Lachnospiraceae, Prevotellaceae, Paraprevotellaceae, Bacteroides, Faecalibacterium. PBC Increase: Enterobacteriaceae such as E. coli, K. pneumoniae, and E. cloacae. Decrease: Faecalibacterium, Bacteroides, Sutterella and Oscillospira spp. AIH Increase: Veillonella Decrease: anaerobes such as Faecalibacterium, Bifidobacterium |
| End stage chronic liver disease | Increase: Veillonillaceaem, Porphyromonadaceae, Streptococcaceae, Enterobacteriaceae, Candida Decrease: Lachnospiraceae and Ruminococcaceae |
| Hepatic encephalopathy | Increase: EnterobacteriasDecrease: Bacteroides |
| Hepatocellular carcinoma | Increase: Bacteroides, Oscillospira, Enterococcus, acteroides, Clostridium, Paraprevotella and Ruminococcaceae Decrease: Clostridium XI y XIVa, Bifidobacterium, Alphaprotreobacteria and Akkermansia |
| ACLF | Increase: Streptococcaceae, Enterobacteriaceae, Alcaligenaceae y Porphyromonadaceae, LachnospiraceaeDecrease: Bacteroidetes, Proteobacteria, Firmicutes |
MAFLD, metabolic associated fatty liver disease; ALD, alcoholic liver disease; PSC, primary sclerosing cholangitis; PBC, primary biliary cholangitis; AIH, autoimmune hepatitis; ACLF, acute on chronic liver failure.
Facts and fictions about gut microbiota
The gut is called “the second brain” because it is wired similarly to the brain, with neurons and neurotransmitters. In many studies, the relationship between the microbiota and multiple diseases has been demonstrated, as well as its participation in their pathophysiology, such as cancer, metabolic disorders, autoimmune inflammatory processes and neuropsychiatric diseases (70,71). Current data on the role of the gut microbiota in CLD is constantly evolving and is not fully understood. The gut microbiota has been shown to play a key role in promoting health. Commensal bacteria produce important SCFAs that stimulate the regeneration of enterocytes, they also promote the preservation of the TJ proteins, preserving the integrity of the intestinal barrier. When the balance of commensal bacteria is disturbed, bacteria such as Proteobacteria increases, promoting the secretion of intestinal inflammatory cytokines. The increased intestinal permeability and dysbiosis cause the continuous passage of PAMPS, such as LPS, bacteria and inflammatory cytokines to the liver, activating Kupfer Cells TLR that activates the innate immune response. In ALD, beyond these facts, acetaldehyde, a metabolite of alcohol metabolism, has been shown to disrupt TJ proteins, reduce intestinal motility, and cause intestinal bacterial overgrowth with higher proportions of Bifidobacterium, Streptococci, and Enterobacteria. Enterococcus faecalis in ALD is highly correlated with the severity of the disease. Similar facts in intestinal permeability have been shown in patients with MAFLD. MAFLD patients have an increase in Escherichia coli associated with susceptibility to leaky gut. In autoimmune liver disease, there has also been demonstrated comparable findings of dysbiosis and leaky gut. In PSC, Klebsiella pneumonia have been shown to alter intestinal integrity inducing pore formation in the epithelial layer. PSC patients also have increase concentration of lactobacillus gasseri that induce inflammatory injury (69). In autoimmune hepatitis, there is an increase in Veillonella disbar, which has been related to the severity of the disease. In PBC an increase in Klebsiella pneumonia in stool has been documented, producing microbiome alterations and increasing intestinal permeability (45). In cirrhosis, it has been described several characteristics in gut microbiota that predispose to the development of cirrhosis complications, among them Lactobacillae enrichment have been found to be associated with enhance neuroinflammation and ammonia levels (72). Another important fact is the relationship of gut microbiota and BA. Alteration in microbiota alter the balance of primary and secondary BA, provoking a disturbance in FXR activation, BA enterohepatic circulation and BA production, causing an increase in metabolic stress and host immune response activation (45). For trying to explain all these effects of gut microbiota within the host, several criteria have been proposed; among them, Hill’s inference criteria that are applicable to several pathologies related to the intestinal microbiota and Koch’s criteria. Koch’s Criteria explain that commensal bacteria may colonize a host this supports the fact of the effectiveness of fecal microbiota transplant for the treatment of several diseases (73,74).
In the other hand, a lot of facts have not yet been proven or remain unknown; among them, the role of probiotic treatment in liver diseases, which could improve dysbiosis and reduce intestinal permeability (75). We are far away for fully understanding the effect of microbiota in host metabolism and liver diseases, therefore additional knowledge of this important field will help us to better understand liver disease development and progression, to propose a better target treatment to our patients. As shown in Table 4.
Table 4
| Strong evidence |
| 1. Colonization by the microbiota of all surfaces begins from birth |
| 2. Environmental factors are the most important for the differentiation of the microbiota |
| 3. There is variability in the conformation of the microbiota between individuals |
| 4. The microbiota plays an important role in the pathogenesis of diseases such as irritable bowel syndrome, ulcerative colitis, Crohn’s disease, liver cirrhosis |
| 5. The microbiota has metabolic, inflammatory and endocrinological functions |
| Medium evidence |
| 1. The microbiota can also influence and change the effectivity of vaccines and it can impact the immune response |
| 2. There are different hypotheses of the influence of the intestinal microbiota on colorectal cancer, impacting on its development and treatment |
| 3. Probiotics may modify Composition and restore “Healthy Microbiota” |
| Weak evidence |
| 1. It is controversial whether dysbiosis is the etiological factor of various diseases or is only a cofactor |
| 2. There is not enough information on the role played by the rest of the components of the microbiota on health (fungi, viruses) |
Conclusions
The gut microbiota plays a crucial role in the pathogenesis and progression of liver diseases. Microbiota alterations have been characterized in several CLD s and, although it is not fully understood, the effect of dysbiosis on the progression of CLD has been described, providing a strong rational to deeply explore the importance of the alteration of the microbiota in the development and progression of CLD. Microbiota taxa could represent important biomarkers and could become important therapeutic targets of patients with CLD.
There is still much to investigate regarding the different molecular mechanisms that link the intestinal microbiota and its complex microbiota-liver-brain axis, this may lead to the development of different treatments for liver diseases where it has been documented that the microbiota plays an important role.
Acknowledgments
Funding: None.
Footnote
Provenance and Peer Review: This article was commissioned by the Guest Editor (Ralf Weiskirchen) for the series “The Pathogenesis of Hepatic Fibrosis: Basic Facts and Clinical Challenges” published in Digestive Medicine Research. The article has undergone external peer review.
Reporting Checklist: The authors have completed the Narrative Review reporting checklist. Available at https://dmr.amegroups.com/article/view/10.21037/dmr-21-85/rc
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dmr.amegroups.com/article/view/10.21037/dmr-21-85/coif). The series “The Pathogenesis of Hepatic Fibrosis: Basic Facts and Clinical Challenges” was commissioned by the editorial office without any funding or sponsorship. The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Boursier J, Diehl AM. Nonalcoholic Fatty Liver Disease and the Gut Microbiome. Clin Liver Dis 2016;20:263-75. [Crossref] [PubMed]
- Ramírez-Pérez O, Cruz-Ramón V, Chinchilla-López P, et al. The Role of the Gut Microbiota in Bile Acid Metabolism. Ann Hepatol 2017;16:S21-6. [Crossref] [PubMed]
- Jandhyala SM, Talukdar R, Subramanyam C, et al. Role of the normal gut microbiota. World J Gastroenterol 2015;21:8787-803. [Crossref] [PubMed]
- Morais LH, Schreiber HL 4th, Mazmanian SK. The gut microbiota-brain axis in behaviour and brain disorders. Nat Rev Microbiol 2021;19:241-55. [Crossref] [PubMed]
- Fouts DE, Torralba M, Nelson KE, et al. Bacterial translocation and changes in the intestinal microbiome in mouse models of liver disease. J Hepatol 2012;56:1283-92. [Crossref] [PubMed]
- Brenner DA, Paik YH, Schnabl B. Role of Gut Microbiota in Liver Disease. J Clin Gastroenterol 2015;49:S25-7. [Crossref] [PubMed]
- Hernández-Ceballos W, Cordova-Gallardo J, Mendez-Sanchez N. Gut Microbiota in Metabolic-associated Fatty Liver Disease and in Other Chronic Metabolic Diseases. J Clin Transl Hepatol 2021;9:227-38. [Crossref] [PubMed]
- Barton W, Shanahan F, Cotter PD, et al. The metabolic role of the microbiota. Clin Liver Dis (Hoboken) 2015;5:91-3. [Crossref] [PubMed]
- Aron-Wisnewsky J, Vigliotti C, Witjes J, et al. Gut microbiota and human NAFLD: disentangling microbial signatures from metabolic disorders. Nat Rev Gastroenterol Hepatol 2020;17:279-97. [Crossref] [PubMed]
- Schenk M, Mueller C. The mucosal immune system at the gastrointestinal barrier. Best Pract Res Clin Gastroenterol 2008;22:391-409. [Crossref] [PubMed]
- Shi N, Li N, Duan X, et al. Interaction between the gut microbiome and mucosal immune system. Mil Med Res 2017;4:14. [Crossref] [PubMed]
- Sellmann C, Priebs J, Landmann M, et al. Diets rich in fructose, fat or fructose and fat alter intestinal barrier function and lead to the development of nonalcoholic fatty liver disease over time. J Nutr Biochem 2015;26:1183-92. [Crossref] [PubMed]
- Albillos A, de Gottardi A, Rescigno M. The gut-liver axis in liver disease: Pathophysiological basis for therapy. J Hepatol 2020;72:558-77. [Crossref] [PubMed]
- Hansson GC, Johansson ME. The inner of the two Muc2 mucin-dependent mucus layers in colon is devoid of bacteria. Gut Microbes 2010;1:51-4. [Crossref] [PubMed]
- Pilewski JM, Frizzell RA. Role of CFTR in airway disease. Physiol Rev 1999;79:S215-55. [Crossref] [PubMed]
- Hermiston ML, Gordon JI. Inflammatory bowel disease and adenomas in mice expressing a dominant negative N-cadherin. Science 1995;270:1203-7. [Crossref] [PubMed]
- Rattan P, Minacapelli CD, Rustgi V. The Microbiome and Hepatocellular Carcinoma. Liver Transpl 2020;26:1316-27. [Crossref] [PubMed]
- Pathak P, Xie C, Nichols RG, et al. Intestine farnesoid X receptor agonist and the gut microbiota activate G-protein bile acid receptor-1 signaling to improve metabolism. Hepatology 2018;68:1574-88. [Crossref] [PubMed]
- Albhaisi SAM, Bajaj JS, Sanyal AJ. Role of gut microbiota in liver disease. Am J Physiol Gastrointest Liver Physiol 2020;318:G84-98. [Crossref] [PubMed]
- Chassaing B, Etienne-Mesmin L, Gewirtz AT. Microbiota-liver axis in hepatic disease. Hepatology 2014;59:328-39. [Crossref] [PubMed]
- Ohtani N, Kawada N. Role of the Gut-Liver Axis in Liver Inflammation, Fibrosis, and Cancer: A Special Focus on the Gut Microbiota Relationship. Hepatol Commun 2019;3:456-70. [Crossref] [PubMed]
- Goel A, Gupta M, Aggarwal R. Gut microbiota and liver disease. J Gastroenterol Hepatol 2014;29:1139-48. [Crossref] [PubMed]
- Gérard C, Vidal H. Impact of Gut Microbiota on Host Glycemic Control. Front Endocrinol (Lausanne) 2019;10:29. [Crossref] [PubMed]
- Sumara G, Sumara O, Kim JK, et al. Gut-derived serotonin is a multifunctional determinant to fasting adaptation. Cell Metab 2012;16:588-600. [Crossref] [PubMed]
- Cunningham AL, Stephens JW, Harris DA. Gut microbiota influence in type 2 diabetes mellitus (T2DM). Gut Pathog 2021;13:50. [Crossref] [PubMed]
- Zhao L, Zhang F, Ding X, et al. Gut bacteria selectively promoted by dietary fibers alleviate type 2 diabetes. Science 2018;359:1151-6. [Crossref] [PubMed]
- Ghosh TS, Rampelli S, Jeffery IB, et al. Mediterranean diet intervention alters the gut microbiome in older people reducing frailty and improving health status: the NU-AGE 1-year dietary intervention across five European countries. Gut 2020;69:1218-28. [Crossref] [PubMed]
- Vatanen T, Franzosa EA, Schwager R, et al. The human gut microbiome in early-onset type 1 diabetes from the TEDDY study. Nature 2018;562:589-94. [Crossref] [PubMed]
- Stewart CJ, Ajami NJ, O'Brien JL, et al. Temporal development of the gut microbiome in early childhood from the TEDDY study. Nature 2018;562:583-8. [Crossref] [PubMed]
- Stepanova M, Younossi ZM. Independent association between nonalcoholic fatty liver disease and cardiovascular disease in the US population. Clin Gastroenterol Hepatol 2012;10:646-50. [Crossref] [PubMed]
- Witkowski M, Weeks TL, Hazen SL. Gut Microbiota and Cardiovascular Disease. Circ Res 2020;127:553-70. [Crossref] [PubMed]
- Trøseid M, Andersen GØ, Broch K, et al. The gut microbiome in coronary artery disease and heart failure: Current knowledge and future directions. EBioMedicine 2020;52:102649. [Crossref] [PubMed]
- Novakovic M, Rout A, Kingsley T, et al. Role of gut microbiota in cardiovascular diseases. World J Cardiol 2020;12:110-22. [Crossref] [PubMed]
- Al Bander Z, Nitert MD, Mousa A, et al. The Gut Microbiota and Inflammation: An Overview. Int J Environ Res Public Health 2020;17:7618. [Crossref] [PubMed]
- Ghosh SS, Wang J, Yannie PJ, et al. Intestinal Barrier Dysfunction, LPS Translocation, and Disease Development. J Endocr Soc 2020;4:bvz039. [Crossref] [PubMed]
- Adolph TE, Grander C, Moschen AR, et al. Liver-Microbiome Axis in Health and Disease. Trends Immunol 2018;39:712-23. [Crossref] [PubMed]
- Hartmann P, Seebauer CT, Schnabl B. Alcoholic liver disease: the gut microbiome and liver cross talk. Alcohol Clin Exp Res 2015;39:763-75. [Crossref] [PubMed]
- Zhong X, Cui P, Jiang J, et al. Streptococcus, the Predominant Bacterium to Predict the Severity of Liver Injury in Alcoholic Liver Disease. Front Cell Infect Microbiol 2021;11:649060. [Crossref] [PubMed]
- Kim SS, Eun JW, Cho HJ, et al. Microbiome as a potential diagnostic and predictive biomarker in severe alcoholic hepatitis. Aliment Pharmacol Ther 2021;53:540-51. [PubMed]
- Yoshioka Y, Hashimoto E, Yatsuji S, et al. Nonalcoholic steatohepatitis: cirrhosis, hepatocellular carcinoma, and burnt-out NASH. J Gastroenterol 2004;39:1215-8. [Crossref] [PubMed]
- Thomas LV, Ockhuizen T, Suzuki K. Exploring the influence of the gut microbiota and probiotics on health: a symposium report. Br J Nutr 2014;112:S1-18. [Crossref] [PubMed]
- Miele L, Valenza V, La Torre G, et al. Increased intestinal permeability and tight junction alterations in nonalcoholic fatty liver disease. Hepatology 2009;49:1877-87. [Crossref] [PubMed]
- Henao-Mejia J, Elinav E, Jin C, et al. Inflammasome-mediated dysbiosis regulates progression of NAFLD and obesity. Nature 2012;482:179-85. [Crossref] [PubMed]
- Chen X, Zhang Z, Li H, et al. Endogenous ethanol produced by intestinal bacteria induces mitochondrial dysfunction in non-alcoholic fatty liver disease. J Gastroenterol Hepatol 2020;35:2009-19. [Crossref] [PubMed]
- Wei Y, Li Y, Yan L, et al. Alterations of gut microbiome in autoimmune hepatitis. Gut 2020;69:569-77. [Crossref] [PubMed]
- Liwinski T, Casar C, Ruehlemann MC, et al. A disease-specific decline of the relative abundance of Bifidobacterium in patients with autoimmune hepatitis. Aliment Pharmacol Ther 2020;51:1417-28. [Crossref] [PubMed]
- Tang R, Wei Y, Li Y, et al. Gut microbial profile is altered in primary biliary cholangitis and partially restored after UDCA therapy. Gut 2018;67:534-41. [Crossref] [PubMed]
- Blesl A, Stadlbauer V. The Gut-Liver Axis in Cholestatic Liver Diseases. Nutrients 2021;13:1018. [Crossref] [PubMed]
- Little R, Wine E, Kamath BM, et al. Gut microbiome in primary sclerosing cholangitis: A review. World J Gastroenterol 2020;26:2768-80. [Crossref] [PubMed]
- Sabino J, Vieira-Silva S, Machiels K, et al. Primary sclerosing cholangitis is characterised by intestinal dysbiosis independent from IBD. Gut 2016;65:1681-9. [Crossref] [PubMed]
- Iwasawa K, Suda W, Tsunoda T, et al. Characterisation of the faecal microbiota in Japanese patients with paediatric-onset primary sclerosing cholangitis. Gut 2017;66:1344-6. [Crossref] [PubMed]
- Rühlemann M, Liwinski T, Heinsen FA, et al. Consistent alterations in faecal microbiomes of patients with primary sclerosing cholangitis independent of associated colitis. Aliment Pharmacol Ther 2019;50:580-9. [Crossref] [PubMed]
- Giannelli V, Di Gregorio V, Iebba V, et al. Microbiota and the gut-liver axis: bacterial translocation, inflammation and infection in cirrhosis. World J Gastroenterol 2014;20:16795-810. [Crossref] [PubMed]
- Horvath A, Rainer F, Bashir M, et al. Biomarkers for oralization during long-term proton pump inhibitor therapy predict survival in cirrhosis. Sci Rep 2019;9:12000. [Crossref] [PubMed]
- Betrapally NS, Gillevet PM, Bajaj JS. Gut microbiome and liver disease. Transl Res 2017;179:49-59. [Crossref] [PubMed]
- Qin N, Yang F, Li A, et al. Alterations of the human gut microbiome in liver cirrhosis. Nature 2014;513:59-64. [Crossref] [PubMed]
- Bajaj JS, Salzman NH, Acharya C, et al. Fecal Microbial Transplant Capsules Are Safe in Hepatic Encephalopathy: A Phase 1, Randomized, Placebo-Controlled Trial. Hepatology 2019;70:1690-703. [Crossref] [PubMed]
- Chen Z, Ruan J, Li D, et al. The Role of Intestinal Bacteria and Gut-Brain Axis in Hepatic Encephalopathy. Front Cell Infect Microbiol 2021;10:595759. [Crossref] [PubMed]
- Zhai S, Zhu L, Qin S, et al. Effect of lactulose intervention on gut microbiota and short chain fatty acid composition of C57BL/6J mice. Microbiologyopen 2018;7:e00612. [Crossref] [PubMed]
- Ponziani FR, Scaldaferri F, Petito V, et al. The Role of Antibiotics in Gut Microbiota Modulation: The Eubiotic Effects of Rifaximin. Dig Dis 2016;34:269-78. [Crossref] [PubMed]
- Horvath A, Leber B, Schmerboeck B, et al. Randomised clinical trial: the effects of a multispecies probiotic vs. placebo on innate immune function, bacterial translocation and gut permeability in patients with cirrhosis. Aliment Pharmacol Ther 2016;44:926-35. [Crossref] [PubMed]
- Dhiman RK. Gut microbiota and hepatic encephalopathy. Metab Brain Dis 2013;28:321-6. [Crossref] [PubMed]
- Lapidot Y, Amir A, Nosenko R, et al. Alterations in the Gut Microbiome in the Progression of Cirrhosis to Hepatocellular Carcinoma. mSystems 2020;5:00153-20. [Crossref] [PubMed]
- Nava GM, Stappenbeck TS. Diversity of the autochthonous colonic microbiota. Gut Microbes 2011;2:99-104. [Crossref] [PubMed]
- Yoshimoto S, Loo TM, Atarashi K, et al. Obesity-induced gut microbial metabolite promotes liver cancer through senescence secretome. Nature 2013;499:97-101. [Crossref] [PubMed]
- Chen Y, Guo J, Qian G, et al. Gut dysbiosis in acute-on-chronic liver failure and its predictive value for mortality. J Gastroenterol Hepatol 2015;30:1429-37. [Crossref] [PubMed]
- Trebicka J, Bork P, Krag A, et al. Utilizing the gut microbiome in decompensated cirrhosis and acute-on-chronic liver failure. Nat Rev Gastroenterol Hepatol 2021;18:167-80. [Crossref] [PubMed]
- Simon TG, Chan AT, Huttenhower C. Microbiome Biomarkers: One Step Closer in NAFLD Cirrhosis. Hepatology 2021;73:2063-6. [Crossref] [PubMed]
- Baratta F, Pastori D, Angelico F, et al. Nonalcoholic Fatty Liver Disease and Fibrosis Associated With Increased Risk of Cardiovascular Events in a Prospective Study. Clin Gastroenterol Hepatol 2020;18:2324-2331.e4. [Crossref] [PubMed]
- Cani PD. Human gut microbiome: hopes, threats and promises. Gut 2018;67:1716-25. [Crossref] [PubMed]
- Feehley T, Plunkett CH, Bao R, et al. Healthy infants harbor intestinal bacteria that protect against food allergy. Nat Med 2019;25:448-53. [Crossref] [PubMed]
- Chopyk DM, Grakoui A. Contribution of the Intestinal Microbiome and Gut Barrier to Hepatic Disorders. Gastroenterology 2020;159:849-63. [Crossref] [PubMed]
- Relman DA. Thinking about the microbiome as a causal factor in human health and disease: philosophical and experimental considerations. Curr Opin Microbiol 2020;54:119-26. [Crossref] [PubMed]
- Neville BA, Forster SC, Lawley TD. Commensal Koch's postulates: establishing causation in human microbiota research. Curr Opin Microbiol 2018;42:47-52. [Crossref] [PubMed]
- Hemarajata P, Versalovic J. Effects of probiotics on gut microbiota: mechanisms of intestinal immunomodulation and neuromodulation. Therap Adv Gastroenterol 2013;6:39-51. [Crossref] [PubMed]
- Jovel J, Dieleman LA, Kao D, et al. The Human Gut Microbiome in Health and Disease. In: Nagarajan M. editor. Metagenomics: Perspectives, Methods, and Applications. Elsevier, 2018:197-213.
- Healey GR, Murphy R, Brough L, et al. Interindividual variability in gut microbiota and host response to dietary interventions. Nutr Rev 2017;75:1059-80. [Crossref] [PubMed]
- Guinane CM, Cotter PD. Role of the gut microbiota in health and chronic gastrointestinal disease: understanding a hidden metabolic organ. Therap Adv Gastroenterol 2013;6:295-308. [Crossref] [PubMed]
- Ciabattini A, Olivieri R, Lazzeri E, et al. Role of the Microbiota in the Modulation of Vaccine Immune Responses. Front Microbiol 2019;10:1305. [Crossref] [PubMed]
- Cheng Y, Ling Z, Li L. The Intestinal Microbiota and Colorectal Cancer. Front Immunol 2020;11:615056. [Crossref] [PubMed]
Cite this article as: Valentin-Cortez FJ, Córdova-Gallardo J, Méndez-Sánchez N. Narrative review of gut microbiota and liver diseases: facts and fictions. Dig Med Res 2022;5:16.


